Carbapenemase genes in clinical and environmental isolates of Acinetobacter spp. from Quito, Ecuador
- Published
- Accepted
- Received
- Academic Editor
- Joseph Gillespie
- Subject Areas
- Microbiology, Public Health
- Keywords
- Carbapenemases, Oxacilinases, Clinical isolates, River isolates, Quito, Acinetobacter, Ecuador, OXA-143, OXA-51, Carbapenem resistance
- Copyright
- © 2024 Sotomayor et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2024. Carbapenemase genes in clinical and environmental isolates of Acinetobacter spp. from Quito, Ecuador. PeerJ 12:e17199 https://doi.org/10.7717/peerj.17199
Abstract
Carbapenem-resistant Acinetobacter spp. is associated with nosocomial infections in intensive care unit patients, resulting in high mortality. Although Acinetobacter spp. represent a serious public health problem worldwide, there are a few studies related to the presence of carbapenemases in health care facilities and other environmental settings in Ecuador. The main aim of this study was to characterize the carbapenem-resistant Acinetobacter spp. isolates obtained from four hospitals (52) and from five rivers (27) close to Quito. We used the disc diffusion and EDTA sinergy tests to determine the antimicrobial susceptibility and the production of metallo β-lactamases, respectively. We carried out a multiplex PCR of gyrB gene and the sequencing of partial rpoB gene to bacterial species identification. We performed molecular screening of nine carbapenem-resistant genes (blaSPM, blaSIM, blaGIM, blaGES, blaOXA-23, blaOXA-24, blaOXA-51, blaOXA-58, and blaOXA-143) by multiplex PCR, followed by identification using sequencing of blaOXA genes. Our findings showed that carbapenem-resistant A. baumannii were the main species found in health care facilities and rivers. Most of the clinical isolates came from respiratory tract samples and harbored blaOXA-23, blaOXA-366, blaOXA-72, blaOXA-65, blaOXA-70, and blaOXA-143-like genes. The river isolates harbored only the blaOXA-51 and probably blaOXA-259 genes. We concluded that the most predominant type of carbapenem genes among isolates were both blaOXA-23 and blaOXA-65 among A. baumannii clinical isolates.
Introduction
Acinetobacter species are aerobic, Gram-negative pleomorphic, and non-lactose fermenting bacilli (Murray, Rosenthal & Pfaller, 2021) and they are ubiquitous in nature, including human skin (Almasaudi, 2016). In the past two decades, multidrug-resistant Acinetobacter spp. have become an important cause of health care-associated infections (HAIs), contributing to increase mortality rates in intensive care units (ICUs) (Castanheira, Mendes & Gales, 2023). Of the all Acinetobacter species, A. baumannii is predominant in clinical settings (Evans, Hamouda & Amyes, 2013; Silva et al., 2022) although non-baumannii species are also considered clinically important (Al Atrouni et al., 2016a). The World Health Organization (2017) elevated A. baumannii to the top of the list of critical pathogens. In Ecuador, A. baumannii was the main bacterium associated with HAIs in the ICU in 2010 and 2011 (Jiménez, 2013).
Acinetobacter species have high carbapenem resistance by the acquisition of genetic determinants of resistance through horizontal gene transfer (Castanheira, Mendes & Gales, 2023). The environment plays a key role in the spread of resistance to clinically relevant antibiotics (Singer et al., 2016); the spread of antibiotic resistance genes in river systems is a potential threat to public health (Wang et al., 2018). Environmental strains harbor antibiotic resistance mechanisms, serving as reservoirs for resistant elements and some of them could cause HAIs (Finley et al., 2013; Al Atrouni et al., 2016a). Horizontal gene transfer plays an important role in the transmission antibiotic resistance between non-pathogens and human pathogens (Wang et al., 2018). Worldwide, A. baumannii has increased the carbapenem resistance in around 90% in the last decade (Rosenthal et al., 2021). In Latin America, Acinetobacter spp. resistance to imipenem and meropenem varied from 8% to 89% (Pan American Health Organization, 2021). In 2010, Ecuador reported that 54% of A. baumannii strains were resistant to meropenem and 51% to imipenem (Labarca et al., 2016); however, the percentage of carbapenem resistant increased to 70% according to the Antimicrobial Resistance Surveillance Network of Ecuador (Satán et al., 2023).
The main mechanism of carbapenem resistance in Acinetobacter species is the production of serin-β-lactamases and metallo-β-lactamases enzymes (Castanheira, Mendes & Gales, 2023). In South America, the most common enzymes are oxacillinases (OXA) encoded by blaOXA-23, blaOXA-72, blaOXA-51, blaOXA-58, blaOXA-235, and blaOXA-143 genes (Escandón-Vargas et al., 2017; García-Betancur et al., 2021). There is scarce information about carbapenem-resistant strains from clinical and environmental settings in Ecuador. In the country, studies reported the presence of blaNDM-1, blaOXA-23, and blaOXA-72 genes in A. baumannii clinical isolates (Nuñez et al., 2016; Villacís et al., 2019). Thus, the main aim of this study was to determine the presence of carbapenem-resistant clinical and river Acinetobacter spp.
Materials and Methods
Clinical isolates
The National Reference Laboratory for Antimicrobial Resistance provided us a total of 52 Acinetobacter spp. clinical isolates recovered through the surveillance network from January to December 2015. As part of the standard procedure, the Reference Laboratory get back the Acinetobacter isolates from four different hospitals: Hospital Pediátrico Baca Ortiz (H1, n = 42), Hospital Pablo Arturo Suárez (H2, n = 5), Hospital del Sur Enrique Garcés (H3, n = 3), and Hospital San Francisco de Quito (H4, n = 2) in Quito (Fig. 1). The Reference Laboratory isolated Acinetobacter spp. from the following clinical samples: pleural fluid, cerebrospinal fluid, blood, urine, catheters, secretions, wounds, skin, abscesses, sputum, tracheal aspirates, and unidentified samples recovered from eleven units (ICU, general surgery, traumatology, burn unit, external consultation, neonatal unit, infectious diseases, pediatric unit, internal medicine, emergency, cardiology, and unidentified units). The data from each isolate are detailed in Table S1. The Reference Laboratory previously identified all isolates as A. baumannii based on VITEK 2 compact system® (Biomerieux, Marcy-I’Étoile, France).
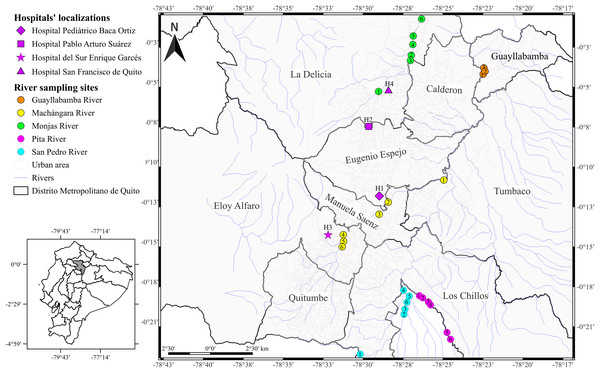
Figure 1: Map of the city of Quito showing sampling locations.
Acinetobacter spp. clinical isolates recovered through the surveillance network from four hospitals in Quito. The hospitals’ localizations are represented by diamond (H1), square (H2), star (H3), and triangle (H4). The river sampling sites are represented by the circles, which indicates the GPS points where we collected the water sample. Rosa de los Ángeles Bayas-Rea made this map.We inoculated the frozen clinical isolates in sheep blood and MacConkey agars and incubated at 37 °C for 24 h. We used Gram staining, oxidase test, and catalase test to re-confirmed the identity of the isolates. We performed DNA extraction by boiling according to the protocol described by Cuaical et al. (2012); briefly, we boiled an overnight culture diluted in sterile water (500 μl) at 100 °C for 15 min, following centrifugation at 10,000 g for 2 min. We quantified the supernatant by spectrophotometry using the Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA) and stored at −20 °C for subsequent amplification. We confirmed species identification by the amplification of partial RNA polymerase β (rpoB) gene and DNA gyrase subunit B (gyrB) gene according to La Scola et al. (2006) and Higgins et al. (2007, 2010a), respectively. The primers selected are detailed in Table 1.
| Target gene | Sequence primer | Size (bp) | References |
|---|---|---|---|
| gyrB | Sp2F: GTTCTTGATCCGAAATTCTCG | 490 | Higgins et al. (2007) |
| Sp4R: AACGGAGCTTGTCAGGGTTA | |||
| Sp4F: CACGCCGTAAGAGTGCATTA | 294 | ||
| Sp4R: AACGGAGCTTGTCAGGGTTA | |||
| Sp1F: GACAACAGTTATAAGGTTTCAGGTG | 428 | Higgins et al. (2010a) | |
| Sp1R: CCGCTATCTGTATCCGCAGTA | |||
| Sp3F: GATAACAGCTATAAAGTTTCAGGTGGT | 194 | ||
| Sp3R: CAAAAACGTACAGTTGTACCACTGC | |||
| rpoB | Ac696F: TAYCGYAAAGAYTTGAAAGAAG | 350 | La Scola et al. (2006) |
| Ac1093R: CMACACCYTTGTTMCCRTG | |||
| Ac1055F: GTGATAARATGGCBGGTCGT | 450 | ||
| Ac1598R: CGBGCRTGCATYTTGTCRT | |||
| blaSIM | F: TACAAGGGATTCGGCATCG | 570 | Ellington et al. (2007) |
| R: TAAGGCCTGTTCCCATGTG | |||
| blaGES | F: ATGCGCTTCATTCACGCAC | 863 | Opazo et al. (2012) |
| R: AACTCATCCTGAGCACGGAC | |||
| blaSPM | F: AAAATCTGGGTACGCAAACG | 271 | Ellington et al. (2007) |
| R: ACATTATCCGCTGGAACAGG | |||
| BlaGIM | F: TCGACACACCTTGGTCTGAA | 477 | Ellington et al. (2007) |
| R: AACTTCCAACTTTGCCATGC | |||
| blaOXA-23 | F: GATCGGATTGGAGAACCAGA | 501 | Woodford et al. (2006) |
| R: ATTTCTGACCGCATTTCCAT | |||
| blaOXA-24 | F: GGTTAGTTGGCCCCCTTAAA | 246 | Woodford et al. (2006) |
| R: AGTTGAGCGAAAAGGGGATT | |||
| blaOXA-51 | F: TAATGCTTTGATCGGCCTTG | 353 | Woodford et al. (2006) |
| R: TGGATTGCACTTCATCTTGG | |||
| blaOXA-58 | F: AAGTATTGGGGCTTGTGCTG | 599 | Woodford et al. (2006) |
| R: CCCCTCTGCGCTCTACATAC | |||
| blaOXA-143 | F: TTCTGTCAGTGCATGCTCATC | 728 | Opazo-Capurro (2014) |
| R: CAGGCATTCCTTGCTTCATT | |||
| blaOXA-51 | F: CTTATAAGTCATATGAACATTAAAGC | 975 | Heritier, Poirel & Nordmann (2006) |
| R: CTCTATAAAAAGGGATCCGGGCTA | |||
| blaOXA-24 | F: ATGAAAAAATTTATACTTCCTA TATTCAGC | 825 | Jeon et al. (2005) |
| R: TTAAATGATTCCAAGATTTTCTAGC | |||
| blaOXA-23 | F: GATGTGTCATAGTATTCGTCG | 1,058 | Afzal-Shah, Woodford & Livermore (2001) |
| R: TCACAACAACTAAAAGCACTG |
Note:
The primer name, the sequences of primer forward and reverse, the expected size, and reference where the sequences were obtained.
River water collection and bacterial identification
We collected the water samples from five principal rivers: Monjas (MO, n = 6), Machángara (MA, n = 6), San Pedro (S, n = 6), Pita (P, n = 6), and Guayllabamba (G, n = 3) in March and April 2016 (Fig. 1 and Table S1). We chose those rivers for two reasons: (i) the four rivers are the main ones that run through Quito, all of them flow into the Guayllabamba River, and (ii) sewage is released into the city’s rivers without prior treatment. In addition, we chose the number of water collection points based on the accessibility and proximity to the urban center. We collected 100 ml of water samples at each river point and transported to the laboratory with ice packs according to Zhang et al. (2013) with few modifications. Subsequently, we centrifuged the water samples at 1,000 g for 10 min at room temperature, resuspended the pellets with 5 ml of fluid thioglycollate medium, and incubated at 37 °C overnight. We inoculated thirty microliters of cultured sample onto CHROMagar Acinetobacter plates with antimicrobial selective supplement (CR 202; CHROMagar Company, Paris, France) and incubated at 37 °C overnight. We selected all colonies with red pigmentation and identified through morphological and conventional biochemical tests used for the clinical isolates. We identified the positive potential isolates using VITEK-2 compact system® (Biomerieux, Marcy-I’Étoile, France). We confirmed the species identification by the amplification of DNA gyrase subunit B (gyrB) gene according to Higgins et al. (2007, 2010a).
Antimicrobial susceptibility test
We use the disc diffusion method and the EDTA sinergy test to determinate the antimicrobial susceptibility and the production of metallo β-lactamases of clinical and river Acinetobacter spp. isolates (Clinical Laboratory Standards Institute, 2022). We spread a concentration corresponding to 0.5 of the McFarland scale of each isolate on Muller Hinton agar plates and incubated at 37 °C overnight. We included the following eleven antimicrobial susceptibility test discs (OXOID, Waltham, MA, USA): ampicillin/sulbactam (20 μg), ceftazidime (30 μg), ciprofloxacin (10 μg), imipenem (10 μg), meropenem (10 μg), gentamicin (10 μg), amikacin (30 μg), cefepime (30 μg), trimethoprim/sulfamethoxazole (25 μg), piperacillin/tazobactam (110 μg), and tobramycin (30 μg). We interpreted the resistance based on the susceptibility breakpoints of the Clinical Laboratory Standards Institute (2022). To assess the synergistic inhibition, we included an ethylenediaminetetraacetic (EDTA) disc (5 mg) with two carbapenem discs, imipenem (10 mg) and meropenem (10 mg), located at 2 cm from the center of the EDTA disc.
Molecular identification of carbapenems-resistant genes
We performed two different multiplex PCRs to screen the presence of carbapenems-resistant genes including six genes encoding enzymes type serine β-lactamases (blaGES, blaOXA-23, blaOXA-24, blaOXA-51, blaOXA-58, and blaOXA-143) and three genes encoding metallo-β-lactamases (blaSPM, blaSIM, and blaGIM) using specific primers previously described (Ellington et al., 2007; Woodford et al., 2006) (Table 1). We achieved a PCR amplification in a final volume of 25 μl containing 1X of Master Mix Green GoTaq® (12.5 μl) (Promega, Madison, WI, USA), 10 μM of forward and reverse primers (0.5 μl), and 50 ng DNA (1 μl). To each PCR assay, we put positive, internal, and negative controls. Both multiplex PCR reaction conditions are detailed in Table 2. We identified all PCR products through electrophoresis in a 2% agarose gel stained with GelstarTM Nucleic Acid Gel Stain 10000 X staining (Lonza, Rockland, ME, USA) and visualized using Gel Doc XR system (Bio-Rad, Hercules, CA, USA).
| Carbapenemase class | Gene | Thermal cycling conditions | Reference | ||||
|---|---|---|---|---|---|---|---|
| Initial denaturation | 35 cycles | Final extention | |||||
| Denaturation | Annealing | Extention | |||||
| Multiplex PCR 1 | blaSPM | 94 °C for 5 min | 94 °C for 30 s | 52.8 °C for 40 s | 72 °C for 50 s | 72 °C for 10 min | Ellington et al. (2007) |
| blaGES | |||||||
| blaSIM | |||||||
| blaGIM | |||||||
| Multiplex PCR 2 | blaOXA-23 | 94 °C for 5 min | 94 °C for 30 s | 53 °C for 30 s | 72 °C for 30 | 72 °C for 10 min | Woodford et al. (2006) |
| blaOXA-24 | |||||||
| blaOXA-51 | |||||||
| blaOXA-58 | |||||||
| blaOXA-143 | |||||||
| Sequencing | blaOXA-51 | 94 °C for 2 min | 94 °C for 1 min | 52 °C for 30 s | 72 °C for 1 min | 72 °C for 10 min | Heritier, Poirel & Nordmann (2006) |
| blaOXA-23 | 94 °C for 5 min | 94 °C for 30 s | 52 °C for 40 s | 72 °C for 50 s | 72 °C for 10 min | Afzal-Shah, Woodford & Livermore (2001) | |
| blaOXA-24 | 94 °C for 5 min | 94 °C for 30 s | 50 °C for 45 s | 72 °C for 1 min | 72 °C for 10 min | Jeon et al. (2005) | |
| blaOXA-143 | 94 °C for 5 min | 94 °C for 30 s | 53 °C for 30 s | 72 °C for 30 | 72 °C for 10 min | Woodford et al. (2006) | |
Note:
The different thermal cycling conditions for various molecular analyses in this study.
We performed a conventional PCR to amplified the entire sequence of the following genes blaOXA-23, blaOXA-24, blaOXA-51, and blaOXA-143 genes using primers previously reported (Heritier, Poirel & Nordmann, 2006; Afzal-Shah, Woodford & Livermore, 2001; Jeon et al., 2005; Woodford et al., 2006) (Table 1), following the conditions detailed in Table 2. We cleaned the PCR products using exonuclease I and shrimp alkaline phosphatase (ExoSAP-IT®). We send both DNA strands of all positive blaOXA genes for sequencing to Macrogen (Seoul, Korea). We aligned manually both forward and reverse sequences for editing and cleaning to obtain a single consensus sequence with the Clustal W program implemented in the MEGA 7 software (Kumar, Stecher & Tamura, 2016). We performed the identification of sequences obtained using a basic local alignment search tool (BLAST) (Altschul et al., 1997). We determined the number of sequences of blaOXA-51 and blaOXA-23 genes with DnaSP v.5.10 software (Rozas et al., 2010).
Results
Identification of Acinetobacter spp.
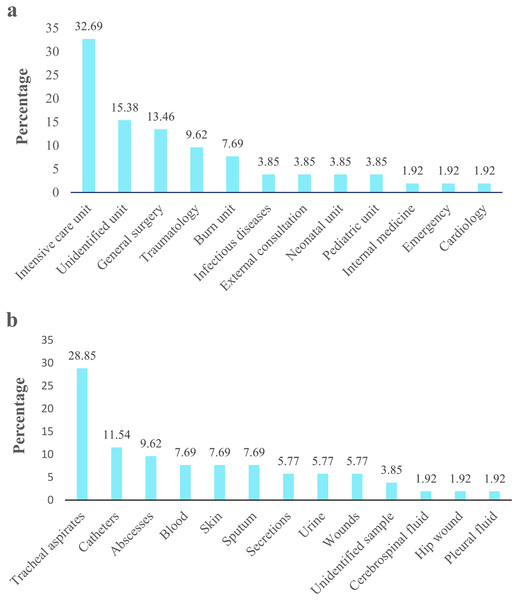
Acinetobacter spp. clinical isolates originated mainly proceeded from pediatric population (77%; 40/52), followed by the adult population (19.2%; 10/52). Isolates derived principally from patients admitted in UCI 32.69% (17/52), followed by unidentified units 15.38% (8/52) and general surgery 13.46% (7/52) (Fig. 2A). According to the clinical specimens, the isolates came from tracheal aspirates 28.85% (15/52), followed by catheters 11.54% (6/52) and abscesses 9.62% (5/52) (Fig. 2B). The majority of clinical isolates were identified as A. baumannii (96.15% 50/52) and A. pittii (3.85%; 2/52) according rpoB gene. Only the identity of A. pittii (3.85%; 2/52) was confirmed with both markers, rpoB and gyrB genes. A. baumannii was found in all type of clinical specimen from all hospital units while A. pittii was found only in tracheal aspirates from UCI. The biochemical and molecular identification of each isolate is detailed in Tables S2 and S3. The nucleotide and amino sequences of partial rpoB gene are detailed in Informations S1 and S2. We submitted all rpoB sequences to GenBank with assigned accession numbers OP796738–OP796781.
Figure 2: Acinetobacter spp. clinical isolates obtained from four hospitals in Quito.
The percentage of Acinetobacter spp. clinical isolates. (A) Isolates obtained from different hospital service units. (B) Isolates recovered from clinical specimens.A total of 27 water samples were collected from five rivers. Twenty-two samples presented suggestive Acinetobacter isolates, four samples (RMO3, RS5, RP4, and RG3) showed no growth, and one sample (RMO4) had isolates not related to Acinetobacter spp. We recovered twenty-six suggestive isolates from San Pedro (2), Machángara (4), Pita (1), and Monjas (3) rivers, of which only 38.5% (10/26) were identified as Acinetobacter spp. based on VITEK 2 compact® (Biomerieux, Marcy-I’Étoile, France) and as A. baumannii based on gyrB gene amplification. The remaining corresponded to other genera (Pseudomonas aeruginosa: 34.6%, Stenotrophomonas maltophila: 19.3%, Burkkolderia cepacia: 3.8%, and Moraxella group: 3.8%). The biochemical and molecular identification of each isolate is detailed in Tables S2 and S3.
Phenotypic antimicrobial susceptibility
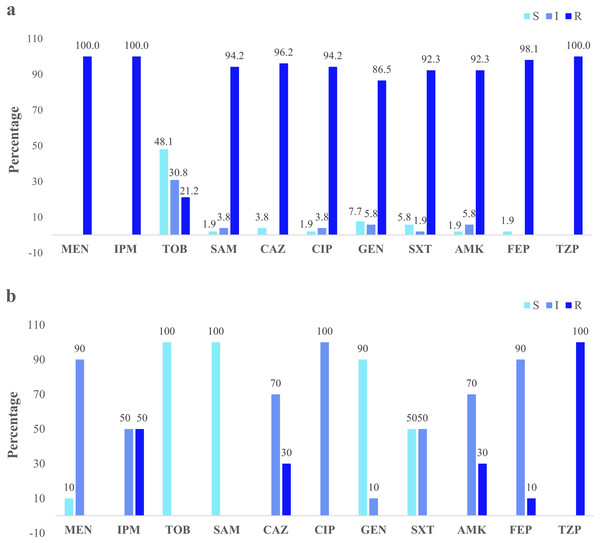
Clinical isolates presented the higher resistance (100%) to the three antibiotics: imipenem, meropenem, and piperacillin/tazobactam. Forty-eight percent were susceptible to tobramycin (Fig. 3A). The river isolates displayed an antimicrobial profile that varied between resistant and intermediate to eight antibiotics tested. Nevertheless, the main resistance was to piperacillin/tazobactam (100%). All of them were susceptible to tobramycin and ampicillin/sulbactam (100%) (Fig. 3B). According to the synergistic inhibition, both river and clinical isolates did not present metallo-β-lactamases enzymes. The antimicrobial susceptibility profile of the clinical and river isolates is detailed in Table S2.
Figure 3: Clinical and environmental isolates susceptibility profile.
The percentages of clinical isolates that were resistant, intermediate, or sensitive to each antibiotic. (A) Clinical isolates susceptibility profile, (B) river isolates susceptibility profile. Acronyms: CIP, ciprofloxacin; GEN, gentamycin; SXT, trimethoprim/sulfamethoxazole; AMK, amikacin; FEP, cefepime; MEN, meropenem; IMP, imipenem; TOB, tobramycin; SAM, ampicillin/sulbactam; CAZ, ceftazidime; TZP, piperacillin/tazobactam; R, resistant; I, intermediate; S, sensitive.Molecular identification of carbapenem-resistance genes
According the screening of carbapenem-resistance genes, clinical isolates presented the following genes: blaSIM, blaSPM, blaGES, blaOXA-23, blaOXA-24, blaOXA-51, and blaOXA-143; whereas, seven river isolates showed blaOXA-51 gene (Table S3). The blaSIM gene, co-harbored with the blaOXA-72 and blaOXA-143 genes, was the least frequently detected (1.9%; 1/52; 15-1175) in A. pittii. The blaSPM gene, co-harbored with the blaOXA-51 and blaOXA-23 genes, was detected in 13.46% (7/52; 15-0122, 15-0591, 15-690, 15-780, 15-795, 15-853, 15-1,138) in A. baumannii. The blaGES gene, co-harbored with the blaOXA-51, blaOXA-23, blaOXA-72, blaOXA-143, and blaSIM genes was present in 11.53% (6/52) in A. baumannii (15-0669, 15-690, 15-853, 15-900) and A. pittii (15-691, 15-1,175). The blaOXA-23 gene, co-harbored with the blaOXA-51 gene, was found in 98.07% (51/52) present in A. baumannii. The blaOXA-24 gene, co-harbored with both blaOXA-23 and blaOXA-51 genes, was present in 9.6% (5/52) in A. baumannii (15-0659, 15-0985, 15-1064, and 15-1367) and in A. pittii (15-0691). The blaOXA-143 gene was the least frequently detected (1.9%; 1/52; 15-1,175); this gene was co-harbored with the blaOXA-24 gene in A. pittii.
Carbapenem-resistance genes sequences analysis
We reported 47 blaOXA-51 sequences: 42 complete sequences of 813 bp and five partial sequences of 340 bp from A. baumannii clinical isolates. Of blaOXA-51 sequences, we identified two genes encoding OXA-65 (n = 44), OXA-70 (n = 2) and one unidentified (MF594746). Forty complete sequences (MF594725–MF594728, MF594730–MF594737, MF594735, MF594739, MF594741–MF594745, OP554146–OP554168) and four partial sequences (MF594729, MF594734, MF594736, and MF594737) shared 100% of nucleotide identity with blaOXA-65 of A. baumannii (GenBank accession number CP033869.1); whereas two entire sequences (MF594740 and MF594747) shared 100% nucleotide identity with blaOXA-70 of A. baumannii (GenBank accession number NG_049811.1).
We found 48 blaOXA-23 sequences: 42 complete sequences of 820 bp and six partial sequences of 471 bp from A. baumannii clinical isolates. Of blaOXA-23 sequences, we identified two genes encoding OXA-23 (n = 38) and OXA-366 (n = 10). Thirty-two entire sequences (MF594756–MF594759, MF594762–MF594767, MF594769, MF594772–MF594777, OP554169, OP554170, OP554172–OP554178, OP554180–OP554182, OP554185–OP554187, OP554189, OP554191–OP554193) and six partial sequences (MF594755, MF594760, MF594761, MF594768, MF594770, and MF594771) shared 100% of nucleotide identity with blaOXA-23 of A. baumannii (GenBank accession number CP110468.1). Ten complete sequences (MF594757, MF59474–MF59476, OP554171, OP554179, OP554183, OP554184, OP554188, and OP554190) shared 100% nucleotide identity with blaOXA-366 of A. baumannii (GenBank accession number NG_049659.1).
We reported five blaOXA-24/40 entire sequences of 817 bp (MF594778, MF594779, MF594781, MF594782, and MF594783) and one partial sequence of 204 bp (MF594780). Of blaOXA-24/40 sequences, we identified one gene encoding OXA-72 present in three A. baumannii and two A. pitti clinical isolates that shared 100% of nucleotide identity with blaOXA-72 of A. baumannii (GenBank accession number MN495626) and also A. pittii (GenBank accession number MN481287.1).
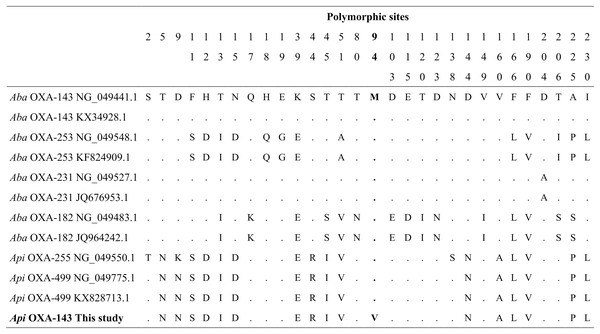
Sequencing of the blaOXA-143 PCR product obtained from A. pitti clinical isolates identified gene encoding OXA-499. Our blaOXA-143 sequence (MF594724) shared 99% nucleotide and amino acid identity with blaOXA-499 of A. pittii (GenBank accession numbers NG_049775.1 and ALM96709.1). The nucleotide sequence presented one specific change in nucleotide 280, first position of the codon (GTG) (ATG), resulting in a valine (V) to methionine (M) change in position 94 (Fig. 4).
Figure 4: Alignment of OXA-143 amino acid sequences.
The polymorphic sites at the amino acid sequence level. The identity of the sequences was compared to the OXA-143 variant (GenBank protein number access WP_063861042.1 or its nucleic number access NG_049441.1). Numbers in the heading row indicate the base pair position of the polymorphic sites. Dots (.) show the identity with the Aba OXA-143. Acronyms: Aba, Acinetobacter baumannii; Api, A. pittii.Sequencing of the blaOXA-51 PCR products obtained from A. baumannii river isolates identified two genes encoding OXA-65 and OXA-259. Four partial sequences (MF594748–MF594751) from three rivers (Pita, Machángara, and San Pedro) shared 100% of nucleotide identity with blaOXA-65 of A. baumannii (GenBank accession number OL961415.1) and three partial sequences (MF594752–MF594754) from Machángara River shared 100% of nucleotide identity with blaOXA-259 of A. baumannii (GenBank accession number CP053098.1).
The nucleotide and amino sequences of blaOXA genes are detailed in the Informations S3 to S10. We submitted all sequences to GenBank with assigned accession numbers MF594724–MF594783, OP554146–OP554193.
Discussion
Acinetobacter spp. in clinical and river isolates
Our findings indicated that Acinetobacter baumannii is the most common species found in clinical isolates from tracheal aspirates from ICU. Similarly, studies reported high rates of baumannii species in ICU samples elsewhere (Qin et al., 2021; Bakhshi et al., 2022). Previous Ecuadorian studies reported high percentages of A. baumannii from respiratory tract samples, bronchial secretions, or tracheal aspirates (Jiménez, 2013; Nuñez et al., 2016; Villacís et al., 2019). On the other hand, we found A. baumannii in the most polluted river environments, Machángara and Monjas rivers (Yungán, 2010; Roldós, 2015). Similar, some studies found these bacteria in river water as well as sewage water (Hrenovic et al., 2016; Kisková et al., 2023). Pathogens of clinical origin as A. baumannii can reach aquatic environments as a result of different human activities, such as sewage water discharge directly into the river without pretreatment (Silva et al., 2016). Acinetobacter spp. survive in wastewater contributing to the spread clinical antibiotic resistance genes in aquatic systems (Kisková et al., 2023; Tobin et al., 2024). Particularly, hospital sewage water is considered a major source of multidrug-resistant pathogenic bacteria and antibiotic residues (Zhang et al., 2013). Contrary, we did not isolate any Acinetobacter spp. from the Guayllabamba River probably due to the distance (2 km), the collection points were far away from the wastewater reservoirs of urban areas and the bacterial concentrations decreases and distribution changes (Zhang et al., 2009).
Carbapenem-resistant Acinetobacter spp.
In our study, the clinical and environmental Acinetobacter isolates showed different susceptibility profiles. Clinical isolates exhibited a high rate of resistance to imipenem and meropenem; while, river isolates displayed a low resistance only to imipenem. In general, clinical isolates have high resistance to both antibiotics (Cortivo et al., 2015). The percentage of resistance found in this study was higher compared with an Ecuadorian study (Jiménez, 2013). River isolates are usually more sensitive compared to clinical isolates. The difference in the patterns of resistance found can be due to various factors such as, biological conditions, virulence factors, epidemiology, or place of origin (Karmostaj, Najar & Salmanian, 2013). In general, it is expected that the antibiotic concentration is lower in aquatic environments than in clinical environments (Zhang et al., 2009; Zhang et al., 2013). In clinical environments, antibiotics (in particular broad-spectrum antibiotics) are supplied continuously, promoting selective pressure in clinical isolates. Following this, the transmission of genetic determinants among clinical isolates could take place through horizontal gene transfer (Zhang et al., 2009; Castanheira, Mendes & Gales, 2023). Nonetheless, the possibility of clonal dissemination as a mean of increasing antibiotic resistance, should not be discarded. This situation generates a faster antibiotic resistance evolution in clinical isolates than in environmental isolates (Zhang et al., 2013). However, the susceptibility results of environmental samples should be treated with caution, as these profiles were based on clinical standards (CLSI) and a small sample size (n = 10).
Carbapenemases genes in Acinetobacter spp.
The high percentage of carbapenem-resistance found in clinical isolates may be increased due to the presence of blaOXA-51, blaOXA-23, blaOXA-72, and blaOXA-143 genes. According to some studies, the association of blaOXA-23 and blaOXA-51 genes as the main cause for carbapenem resistance (i.e., Martínez & Mattar, 2012; Teixeira et al., 2013; Cortivo et al., 2015). The blaOXA-72 gene increases its resistance activity in the presence of imipenem and ampicillin/sulbactam (Kuo et al., 2013). The blaOXA-143 and blaOXA-72 genes in association with intrinsic mechanisms as the alteration of the of the outer membrane proteins promotes a high resistance to meropenem (Mostachio et al., 2012; Sen & Joshi, 2015). The low percentage of carbapenem-resistance found in environmental isolates may be explained by the absence of blaOXA genes. It is necessary the presence of insertion elements to promote overexpression of the blaOXA-51 gene since this gene it generates a weak resistance (Higgins, Zander & Seifert, 2013; Teixeira et al., 2013; Sen & Joshi, 2015). The blaOXA-51 overexpression would be linked to the resistance to imipenem in A. baumannii river isolates (Martínez & Mattar, 2012; Evans, Hamouda & Amyes, 2013). It would be important to know about possible ISAba1 insertions upstream of the gene or alternative genes conferring resistance in the five isolates with resistance phenotype.
We found a high percentage of blaOXA-23 and blaOXA-51 genes in A. baumannii and A. pittii clinical isolates. The high percentage of blaOXA-23 gene found in A. baumannii, is related to its distribution worldwide since a single sequence was found in the study. The blaOXA-23 gene distribution has been associated with a clonal spread (Higgins et al., 2010b; Labarca et al., 2016). In the other hand, blaOXA-51 gene has been reported principally in baumannii species (Lee et al., 2012; Teixeira et al., 2013; Périchon et al., 2014). Since blaOXA-51 gene can be found in the chromosome (Poirel, Naas & Nordmann, 2010), some authors consider it as an intrinsic marker for A. baumannii identification (Higgins et al., 2010b; Périchon et al., 2014). However, the gene may be in transposons and plasmids (Liu et al., 2015; Wang et al., 2015; Sennati et al., 2016). Its dissemination among other Acinetobacter species from different environments is possible, suggesting that the dissemination may not be by clonal strains (See Labarca et al., 2016).
Our findings detected two different blaOXA-51 variants OXA-65 and OXA-70 were circulating in the four hospitals; while, in rivers probably are OXA-65 and OXA-259, suggesting that the genes were under selective pressure. Similarly, previous studies OXA-65 was reported in A. baumannii clone ST79 from Guayaquil (Rodríguez et al., 2016). In Latin America, is one of the variants found in A. baumannii in Brazil, Argentina, Paraguay, Mexico, and Honduras (Rodríguez et al., 2016; Nodari et al., 2020; Graña-Miraglia et al., 2020). There is not report about the presence of OXA-70 neither OXA-259 variant in South America. The information about OXA-70 variant is scare worldwide. OXA-70 has been reported in Hong Kong, Malaysia, Ghana, and Honduras (Alattraqchi et al., 2020; Ayibieke et al., 2020; Brown & Amyes, 2005; Galac et al., 2020). OXA-70 belong to the most diverse collection of class D carbapenemases, subgroup 3 that is the major mechanism of carbapenem resistance in A. baumannii (Brown & Amyes, 2005). OXA-259 variant has been reported in A. baumannii clinical isolates from China, Tanzania, and Thailand (Jia et al., 2019; Moyo et al., 2021; Chukamnerd et al., 2022) as well in bulk tank milk in Germani (Sykes et al., 2023). The variant OXA-259 is located on the chromosome of A. baumannii and constitute an intrinsic oxacillinase genes with low-level carbapenemase activity that naturally occur (Chukamnerd et al., 2022), due there is a not ISAbalike elements (Fedrigo et al., 2022). To the best of our knowledge, this is the first report of the OXA-70 and OXA-259 in A. baumannii in Ecuador.
Our findings showed the presence of blaOXA-366 gene in both species A. baumannii and A. pittii clinical isolates from Ecuador. The information about this variant is scare. GenBank previously reported a sequence of blaOXA-366 in A. baumannii from USA. Whereas the OXA-366 variant has been reported also in A. baumannii clinical isolates in Iran (Bahador et al., 2015). To the best of our knowledge, this is the first report of the OXA-366 in A. baumannii and A. pittii in South America.
The blaOXA-72 gene co-harbored with blaOXA-23 and blaOXA-51 genes was identified in low levels in A. baumannii and A. pittii. This variant was documented previously in A. baumannii clinical isolates from Guayaquil (Nuñez et al., 2016; Rodríguez et al., 2016) and from neighboring countries Colombia, Peru, and Brazil (Saavedra et al., 2014; Saavedra et al., 2017; Werneck et al., 2011; de Sá Cavalcanti et al., 2013; Levy-Blitchtein et al., 2018; Nodari et al., 2020) and in A. pittii from Colombia (Montealegre et al., 2012), Brazil (Brasiliense et al., 2019), European and Asian countries (Al Atrouni et al., 2016b; Bonnin et al., 2014). The presence of this gene in two species may be due to the blaOXA-72 gene spreads through horizontal gene transfer mediated by plasmids (Montealegre et al., 2012; Kuo et al., 2013; Nuñez et al., 2016; Rodríguez et al., 2016). To the best of our knowledge, this is the first report of the blaOXA-72 gene in A. pittii in Ecuador.
This is also the first report of the blaOXA-143 gene in A. pittii clinical isolates from Ecuador. In South America, this gene and its variants (OXA-231, OXA-253, and OXA-255) have been reported in A. baumannii clinical isolates in Brazil (Higgins et al., 2009; Gionco et al., 2012; Girlich et al., 2014), Perú (Levy-Blitchtein et al., 2018), and Colombia (Saavedra et al., 2017). The variant found in this study is very different from OXA-143 variants reported in A. baumannii from Brazil, Honduras, and Korea (Higgins et al., 2009; Zander et al., 2014; Kim et al., 2010). Interestingly, the sequence alignment showed a high similarity between our variant with two OXA-143 variants (OXA-499 and OXA-255) described to A. pittii in Korea (D’Souza et al., 2017) and United States (Zander et al., 2014). In particular, the difference between the variant reported in this study differs by a change in amino acid sequence with OXA-499 reported by D’Souza et al. (2017). To the best of our knowledge, this is the first report of a variant similar to OXA-499 in A. pittii in South America.
Conclusions
Overall, our results showed that the most of the Acinetobacter spp. clinical isolates came from tracheal aspirates from ICU. Our findings showed that carbapenem-resistant A. baumannii were the main species found in the study. We reported the presence of blaOXA-65, blaOXA-70, blaOXA-72, blaOXA-23, blaOXA-366, and blaOXA143-carrying multidrug-resistant genes in baumannii and no baumannii species. We report a new possible variant of OXA-143 circling in different hospitals in Quito and the possible presence of OXA-259 in aquatic environment. The presence of A. baumannii in the rivers increases the concern, since the resistance can be disseminated, polluting water sources, and promoting community acquired outbreaks.
To our knowledge, this is the first study that has characterized the carbapenemases genes in Acinetobacter spp. carbapenem-resistant isolates from four hospitals and five rivers in Quito. No blaOXA-65, blaOXA-70, blaOXA-72, blaOXA-23, blaOXA-366, or blaOXA143-carrying multidrug-resistant baumanii and no baumannii species have been described in Quito so far.
Supplemental Information
Clinical and River isolates data.
All samples correspond to Zone 9, which represents the code for Quito. The table details the hospital code, INSPI isolate code, geographical coordinates age, sex, sample type, and hospital unit of the patients to which each sample corresponds. In addition, the table indicated the river data, id code, geographical coordinates, and river name. Acronyms: NA: not available.
Biochemical results and antibiotic susceptibility profile of Acinetobacter spp.
NA: Not applied, Aba: A. baumannii, Aha: A. haemolyticus, ABC: A. baumannii/calcoaceticus complex, ng/µl: nanograme/microlitre, Y: yes, N: not. Biochemical results are expressed as positive (+) and negative (−). The antibiotic susceptibility profile are expresed as Intermediate (I), Sensistive (S), and Resistance (R).
Molecular results of Acinetobacter spp. obtained by amplification and sequencing of different genes.
A. baumannii was identified by the amplification of both products, 490 bp and 294 bp, and A. pittii with a product of 194 bp of gyrB gene. The species identification was corroborate by sequencing on partial rpo gene. PCR results are expressed for carbapenen resistance genes as positive (+) and negative (−). The table details the GenBank number acccess of sequences obtained in this study.