Fast and exact fixed-radius neighbor search based on sorting
- Published
- Accepted
- Received
- Academic Editor
- Massimiliano Fasi
- Subject Areas
- Algorithms and Analysis of Algorithms, Data Mining and Machine Learning, Data Science, Databases, Neural Networks
- Keywords
- Near neighbor search, Fixed-radius search
- Copyright
- © 2024 Chen and Güttel
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ Computer Science) and either DOI or URL of the article must be cited.
- Cite this article
- 2024. Fast and exact fixed-radius neighbor search based on sorting. PeerJ Computer Science 10:e1929 https://doi.org/10.7717/peerj-cs.1929
Abstract
Fixed-radius near neighbor search is a fundamental data operation that retrieves all data points within a user-specified distance to a query point. There are efficient algorithms that can provide fast approximate query responses, but they often have a very compute-intensive indexing phase and require careful parameter tuning. Therefore, exact brute force and tree-based search methods are still widely used. Here we propose a new fixed-radius near neighbor search method, called SNN, that significantly improves over brute force and tree-based methods in terms of index and query time, provably returns exact results, and requires no parameter tuning. SNN exploits a sorting of the data points by their first principal component to prune the query search space. Further speedup is gained from an efficient implementation using high-level basic linear algebra subprograms (BLAS). We provide theoretical analysis of our method and demonstrate its practical performance when used stand-alone and when applied within the DBSCAN clustering algorithm.
Introduction
This work is concerned with the retrieval of nearest neighbors, a fundamental data operation. Given a data point, this operation aims at finding the most similar data points using a predefined distance function. Nearest neighbor search has many applications in computer science and machine learning, including object recognition (Philbin et al., 2007; Nister & Stewenius, 2006), image descriptor matching (Silpa-Anan & Hartley, 2008), time series indexing (Keogh & Ratanamahatana, 2005; Chakrabarti et al., 2002; Yagoubi et al., 2020), clustering (Ester et al., 1996; Campello, Moulavi & Sander, 2013; Gallego et al., 2018; Alshammari, Stavrakakis & Takatsuka, 2021; Gallego, Rico-Juan & Valero-Mas, 2022; Li et al., 2020), particle simulations (Groß, Köster & Krüger, 2019), molecular modeling (Galvelis & Sugita, 2017), pose estimation (Shakhnarovich, Viola & Darrell, 2003), computational linguistics (Kaminska, Cornelis & Hoste, 2021), and information retrieval (Geng et al., 2008; Wang et al., 2015).
There are two main types of nearest neighbor (NN) search: k-nearest neighbor and fixed-radius near neighbor search. Fixed-radius NN search, also referred to as radius query, aims at identifying all data points within a given distance from a query point; see Bentley (1975b) for a historical review. The most straightforward way of finding nearest neighbors is via a linear search through the whole database, also known as exhaustive or brute force search. Though considered inelegant, it is still widely used, e.g., in combination with GPU acceleration (Garcia, Debreuve & Barlaud, 2008).
Existing NN search approaches can be broadly divided into exact and approximate methods. In many applications, approximate methods are an effective solution for performing fast queries while allowing for a small loss. Well-established approximate NN search techniques include randomized -d trees (Silpa-Anan & Hartley, 2008), hierarchical k-means (Nister & Stewenius, 2006), locality sensitive hashing (Indyk & Motwani, 1998), HNSW (Malkov & Yashunin, 2020), and ScaNN (Guo et al., 2020). Considerable drawbacks of most approximate NN search algorithms are their potentially long indexing time and the need for the tuning of additional hyperparameters such as the trade-off between recall vs index and query time. Furthermore, to the best of our knowledge, all approximate NN methods for which open-source implementations (such as Guo et al., 2020; Bernhardsson, 2023; Dong, Moses & Li, 2011; Malkov & Yashunin, 2020) are available only address the k-nearest neighbor problem, not the fixed-radius problem discussed here.
In this article we introduce a new exact approach to fixed-radius NN search based on sorting, referred to as SNN for short. Some of the appealing properties of SNN are
simplicity: SNN has no hyperparameters except for the necessary search radius
exactness: SNN is guaranteed to return all data points within the search radius
speed: SNN demonstrably outperforms other exact NN search algorithms like, e.g., methods based on tree structures
flexibility: the low indexing time of SNN makes it applicable in an online streaming setting.
The rest of this article is organized as follows. In “Related Work”, we provide a brief review of existing work on NN search. In “Sorting-Based NN Search”, we introduce our sorting-based NN method, detailing its indexing and query phases. “Computational Considerations” contains computational considerations regarding the efficient implementation of SNN and its behavior in floating-point arithmetic. Theoretical performance analysis is provided in “Theoretical Analysis”. “Experimental Evaluation” contains performance comparisons of our algorithm to other state-of-the-art NN methods, as well as an application to DBSCAN clustering. We then conclude in “Conclusions”. (A preprint of this work is available on arXiv (Chen & Güttel, 2023)).
Related work
NN search methods can broadly be classified into approximate or exact methods, depending on whether they return exact or approximate answers to queries (Cayton & Dasgupta, 2007). It is widely accepted that for high-dimensional data there are no exact NN search methods which are asymptotically more efficient than exhaustive search; see, e.g., (Muja, 2013, Chap. 3) and Francis-Landau & Durme (2019). Exact NN methods based on -d tree (Bentley, 1975a; Friedman, Bentley & Finkel, 1977), balltree (Omohundro, 1989), VP-tree (Yianilos, 1993), cover tree (Beygelzimer, Kakade & Langford, 2006), and RP tree (Dasgupta & Sinha, 2013) only perform well on low-dimensional data. This shortcoming is often referred to as the curse of dimensionality (Indyk & Motwani, 1998). However, note that negative asymptotic results do not rule out the possibility of algorithms and implementations that perform significantly (by orders of magnitude) faster than brute force search in practice, even on real-world high-dimensional data sets.
To speedup NN search, modern approaches generally focus on two aspects, namely indexing and sketching. The indexing aims to construct a data structure that prunes the search space for a given query, hopefully resulting in fewer distance computations. Sketching, on the other hand, aims at reducing the cost of each distance computation by using a compressed approximate representation of the data.
The most widely used indexing strategy is space partitioning. Some of the earliest approaches are based on tree structures such as -d tree (Bentley, 1975a; Friedman, Bentley & Finkel, 1977), balltree (Omohundro, 1989), VP-tree (Yianilos, 1993), and cover tree (Beygelzimer, Kakade & Langford, 2006). The tree-based methods are known to become inefficient for high-dimensional data. One of the remedies are randomization (e.g., Dasgupta & Sinha, 2013; Ram & Sinha, 2019) and ensembling (e.g., the FLANN nearest neighbor search tool by Muja & Lowe (2009), which empirically shows competitive performance against approximate NN methods). Another popular space partitioning method is locality-sensitive hashing (LSH); see, e.g., Indyk & Motwani (1998). LSH leverages a set of hash functions from the locality-sensitive hash family and it guarantees that similar queries are hashed into the same buckets with higher probability than less similar ones. This method was originally introduced for the binary Hamming space by Indyk & Motwani (1998), and it was later extended to the Euclidean space (Datar et al., 2004). In Bawa, Condie & Ganesan (2005) a self-tuning index for LSH based similarity search was introduced. A partitioning approach based on neural networks and LSH was proposed in Dong et al. (2020). Another interesting method is GriSPy (Chalela et al., 2021), which performs fixed-radius NN search using regular grid search—to construct a regular grid for the index—with the possibility of working with periodic boundary conditions. This method, however, has high memory demand because the grid computations grow exponentially with the space dimension.
This article focuses on exact fixed-radius NN search. The implementations available in the most widely used scientific computing environments are all based on tree structures, including findNeighborsInRadius in MATLAB (The MathWorks Inc., 2022), NearestNeighbors in scikit-learn (Pedregosa et al., 2011), and spatial in SciPy (Virtanen et al., 2020). This is in contrast to our SNN method introduced below which does not utilise any tree structures.
Sorting-based nn search
Suppose we have data points (represented as column vectors) and . The fixed-radius NN problem consists of finding the subset of data points that is closest to a given query point (may be out-of-sample) with respect to some distance metric. Throughout this article, the vector norm is the Euclidean one, though it also possible to identify nearest neighbors with other distances such as
-
cosine distance: assuming normalized data (with ), the cosine distance is
Hence, the cosine distance can be computed from the Euclidean distance via
-
angular distance: the angular distance between two normalized vectors satisfies
Therefore, closest angle neighbors can be identified via Euclidean distance.
-
maximum inner product similarity (see, e.g., Bachrach et al., 2014): for not necessarily normalized vectors we can consider the transformed data points with and the transformed query point . Then
Since and are independent of the index , we have .
Manhattan distance: since , any points satisfying must necessarily satisfy . Hence, the sorting-based exclusion criterion proposed in section 3.2 to prune the query search space can also be used for the Manhattan distance.
Our algorithm, called SNN, will return the required indices of the nearest neighbors in the Euclidean norm, and can also return the corresponding distances if needed. SNN uses three essential ingredients to obtain its speed. First, a sorting-based exclusion criterion is used to prune the search space for a given query. Second, pre-calculated dot products of the data points allow for a reduction of arithmetic complexity. Third, a reformulation of the distance criterion in terms of matrices (instead of vectors) allows for the use of high-level basic linear algebra subprograms (BLAS, Blackford et al., 2002). In the following, we explain these ingredients in more detail.
Indexing
Before sorting the data, all data points are centered by subtracting the empirical mean value of each dimension:
This operation will not affect the pairwise Euclidean distance between the data points and can be performed in operations, i.e., with linear complexity in . We then compute the first principal component , i.e., the vector along which the data exhibits largest empirical variance. This vector can be computed by a thin singular value decomposition of the tall-skinny data matrix ,
(1) where and have orthonormal columns and is a diagonal matrix such that The principal components are given as the columns of and we require only the first column . The score of a point along is
where denotes the -th canonical unit vector in . In other words, the scores of all points can be read off from the first column of times . The computation of the scores using a thin SVD requires operations and is therefore linear in .
The next (and most important) step is to order all data points by their scores; that is,
so that with each . This sorting will generally require a time complexity of independent of the data dimension . We also precompute the squared-and-halved norm of each data point, for . This is of complexity , i.e., again linear in .
All these computations are done exactly once in the indexing phase and only the scores , the numbers , and the single vector need to be stored. See Algorithm 1 for a summary.
| 1: Input: Data matrix |
| 2: Compute |
| 3: Compute the mean-centered matrix X with rows |
| 4: Compute the singular value decomposition of |
| 5: Compute the sorting keys for |
| 6: Sort data points X such that |
| 7: Compute for |
| 8: Return: μ, X, v1, , |
Query
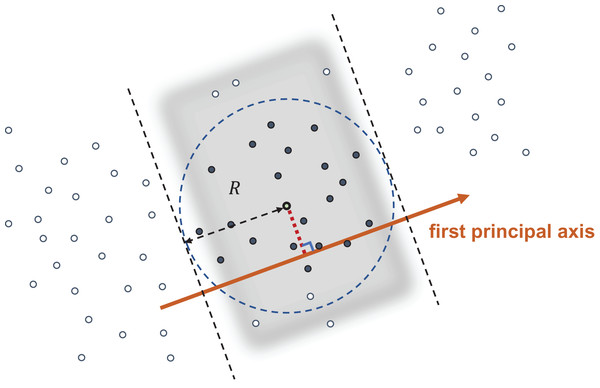
Given a query point and user-specified search radius R, we want to retrieve all data points satisfying . Figure 1 illustrates our approach. We first compute the mean-centered query and the corresponding score . By utilizing the Cauchy–Schwarz inequality, we have
Figure 1: Query with radius R.
The data points in the shaded band have their first principal coordinate within a distance R from the first principal coordinate of the query point, and hence are NN candidates. All data points are sorted so that all candidates have continuous indices.(2)
Since we have sorted the such that , the following statements are true:
As a consequence, we only need to consider candidates whose indices are in and we can determine the smallest subset by finding the largest and smallest satisfying the above statements, respectively. As the are sorted, this can be achieved via binary search in operations. Note that the indices in J are continuous integers, and hence it is memory efficient to access , the submatrix of X whose row indices are in J. This will be important later.
Finally, we filter all data points in the reduced set , retaining only those data points whose distance to the query point is less or equal to R, i.e., points satisfying . The query phase is summarized in Algorithm 2.
| 1: Input: Query vector q; user-specified radius R; output from Algorithm 1 |
| 2: Compute |
| 3: Compute the sorting score of xq, i.e., |
| 4: Select candidate index range J so that for all |
| 5: Compute using the precomputed |
| 6: Return: Points xj with according to Eq. (4) |
Computational considerations
The compute-intensive step of the query procedure is the computation of
(3) for all vectors with indices . Assuming that these vectors have features, one evaluation of Eq. (3) requires floating point operations (flop): flop for the subtractions, flop for the squaring, and flop for the summation. In total, flop are required to compute all |J| squared distances. We can equivalently rewrite (Eq. (3)) as and instead verify the radius condition as
(4)
This form has the advantage that all the squared-and-halved norms ( ) have been precomputed during the indexing phase. Hence, in the query phase, the left-hand side of Eq. (4) can be evaluated for all |J| points using only flop: for the inner products and |J| subtractions.
Merely counting flop, Eq. (4) saves about 1/3 of arithmetic operations over (Eq. (3)). An additional advantage results from the fact that all inner products in Eq. (4) can be computed as using level-2 BLAS matrix-vector multiplication (gemv), resulting in further speedup on modern computing architectures. If multiple query points are given, say , a level-3 BLAS matrix-matrix multiplication (gemm) evaluates in one go, where J is the union of candidates for all query points.
One may be concerned that the computation using Eq. (4) incurs more rounding error than the usual Formula (3). We now prove that this is not the case. First, note that division or multiplication by 2 does not incur rounding error. Using the standard model of floating point arithmetic, we have for any elementary operation , where with the unit roundoff (Higham, 2002, Chap. 1). Suppose we have two vectors and where and denote their respective coordinates. Then computing
in floating point arithmetic amounts to evaluating
Continuing this recursion we arrive at
Assuming and using (Higham, 2002, Lemma 3.1) we have
Hence,
showing that the left-hand side of Eq. (3) can be evaluated with high relative accuracy.
A very similar calculation can be done for the formula
the expression that is used to derive (Eq. (4)). Using the standard result for inner products (Higham, 2002, eq. (3.2))
one readily derives
the same bound on the relative accuracy of floating-point evaluation as obtained for (3).
Theoretical analysis
The efficiency of the SNN query in Algorithm 2 is dependent on the number of pairwise distance computations that are performed in Step 5, depending on the size of the index set |J|. If the index set J is the full , then the algorithm reduces to exhaustive search over the whole dataset , which is undesirable. For the algorithm to be most efficient, |J| would exactly coincide with the indices of data points that satisfy . In practice, the index set J will be somewhere in between these two extremes. Thus, it is natural to ask: How likely is it that , yet ?
First note that, using the singular value decomposition (Eq. (1)) of the data matrix X, we can derive an upper bound on that complements the lower bound (Eq. (2)). Using that , where denotes the th canonical unit vector, and denoting the elements of U by , we have
with Therefore,
(5) and the gap in these inequalities depends on , the second singular value of X. Indeed, if , then all data points lie on a straight line passing through the origin and their distances correspond exactly to the difference in their first principal coordinates. This is a best-case scenario for Algorithm 2 as all candidates , , found in Step 4 are indeed also nearest neighbors. If, on the other hand, is relatively large compared to , the gap in the inequalities (Eq. (5)) becomes large and may be a crude underestimation of the distance .
In order to get a qualitative understanding of how the number of distance computations in Algorithm 2 depends on the various parameters (dimension , singular values of the data matrix, query radius R, etc.), we consider the following model. Let be a large sample of points whose components are normally distributed with zero mean and standard deviation , , respectively. These points describe an elongated “Gaussian blob” in , with the elongation controlled by . In the large data limit the singular values of the data matrix approach and the principal components approach the canonical unit vectors . As a consequence, the principal coordinates follow a standard normal distribution, and hence for any the probability that is given as
On the other hand, the probability that is given by
(6) where F denotes the cumulative distribution function. In this model we can think of the point as a query point, and our aim is to identify all data points within a radius R of this query point.
Since implies that , we have . Hence, the quotient can be interpreted as a conditional probability of a point satisfying given that , i.e.,
Ideally, we would like this quotient be close to , and it is now easy to study the dependence on the various parameters. First note that does not depend on nor , and hence the only effect these two parameters have on P is via the factor in the integrand of . This term corresponds to the probability that the sum of squares of independent Gaussian random variables with mean zero and standard deviation is less or equal to . Hence, and therefore P are monotonically decreasing as or are increasing. This is consistent with intuition: as increases, the elongated point cloud becomes more spherical and hence it gets more difficult to find a direction in which to enumerate (sort) the points naturally. And this problem gets more pronounced in higher dimensions .
We now show that the “efficiency ratio” P converges to as R increases. In other words, the identification of candidate points , , should become relatively more efficient as the query radius R increases. (Here relative is meant in the sense that candidate points become more likely to be fixed-radius nearest neighbors as R increases. Informally, as , all data points are candidates and also nearest neighbors and so the efficiency ratio must be .) First note that for an arbitrarily small there exists a radius such that . Further, there is a such that
To see this, note that the cumulative distribution function F increases monotonically from to as its first argument increases from to . Hence there exists a value T for which for all . Now we just need to find such that
The left-hand side is a quadratic function with roots at , symmetric with respect to the maximum at . Hence choosing such that
or any value larger than that, will be sufficient. Now, setting , we have
Hence, both and come arbitrarily close to as R increases, and so does their quotient .
Experimental evaluation
Our experiments are conducted on a compute server with two Intel Xeon Silver 4114 2.2G processors, 1.5 TB RAM, with operating system Linux Debian 11. All algorithms are forced to run in a single thread with the same settings for fair comparison. We only consider algorithms for which stable Cython or Python implementation are freely available. Our SNN algorithm is implemented in native Python (i.e., no Cython is used), while scikit-learn’s (Pedregosa et al., 2011) -d tree and balltree NN algorithms, and hence also scikit-learn’s DBSCAN method, use Cython for some part of the computation. Numerical values are reported to four significant digits. The code and data to reproduce the experiments in this article can be downloaded from https://github.com/nla-group/snn.
Near neighbor query on synthetic data
We first compare -d tree, balltree, and SNN on synthetically generated data to study their dependence on the data size and the data dimension . We also include two brute force methods, the one in scikit-learn (Pedregosa et al., 2011) (denoted as brute force 1) and another one implemented by us (denoted as brute force 2) which exploits BLAS level-2 (One might say that brute force 2 is equivalent to SNN without index construction and without search space pruning.). The leaf size for scikit-learn’s -d tree and balltree is kept at the default value 40. The data points are obtained by sampling from the uniform distribution on .
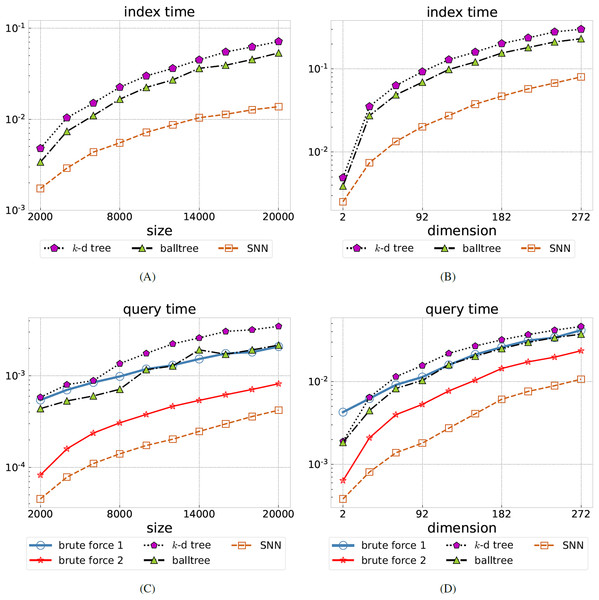
For the first test we vary the number of data points (the index size) from 2,000 to 20,000 in increments of 2,000. The number of features is either or . We then query the nearest neighbors of each data point for varying radius R. The ratio of returned data points relative to the overall number of points is listed in Table 1. As expected, this ratio is approximately independent of . We have chosen the radii R so that a good order-of-magnitude variation in the ratio is obtained, in order to simulate queries with small to large returns. The timings of the index and query phases of the various NN algorithms are shown in Fig. 2 (left). Note that the brute force methods do not require the construction of an index. Among -d tree, balltree, and SNN, our method has the shortest indexing phase. The query time is obtained as an average over all queries, over the two considered dimensions , and over all considered radii R.SNN performs best, with the average query time being between 5 and 9.7 times faster than balltree (the fastest tree-based method). We have verified that for all methods, when run on the same datasets with the same radius parameter, the set of returned points coincide.
| 2,000 | 4,000 | 6,000 | 8,000 | 10,000 | 12,000 | 14,000 | 16,000 | 18,000 | 20,000 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Varying ( ) | 0.1243% | 0.1245% | 0.1232% | 0.1234% | 0.1236% | 0.1236% | 0.1238% | 0.1238% | 0.1232% | 0.1234% | |
| 0.755% | 0.7561% | 0.7506% | 0.7522% | 0.7532% | 0.7508% | 0.7531% | 0.7534% | 0.7511% | 0.751% | ||
| 1.852% | 1.87% | 1.879% | 1.871% | 1.882% | 1.879% | 1.879% | 1.871% | 1.881% | 1.875% | ||
| 3.434% | 3.46% | 3.433% | 3.449% | 3.44% | 3.459% | 3.454% | 3.44% | 3.452% | 3.453% | ||
| 5.487% | 5.42% | 5.443% | 5.456% | 5.438% | 5.454% | 5.442% | 5.432% | 5.449% | 5.417% | ||
| Varying ( ) | 0.01732% | 0.01674% | 0.01818% | 0.01763% | 0.01752% | 0.01717% | 0.01734% | 0.01726% | 0.0175% | 0.0174% | |
| 0.07652% | 0.07361% | 0.07286% | 0.07502% | 0.07777% | 0.07414% | 0.07571% | 0.07737% | 0.07486% | 0.07722% | ||
| 0.2873% | 0.2843% | 0.2912% | 0.2863% | 0.2879% | 0.2857% | 0.2862% | 0.2903% | 0.2888% | 0.2892% | ||
| 0.9608% | 0.9235% | 0.9316% | 0.9184% | 0.9129% | 0.929% | 0.9065% | 0.9195% | 0.9166% | 0.9303% | ||
| 2.514% | 2.624% | 2.623% | 2.544% | 2.526% | 2.511% | 2.542% | 2.558% | 2.584% | 2.562% | ||
| 2 | 32 | 62 | 92 | 122 | 152 | 182 | 212 | 242 | 272 | ||
| Varying ( 10,000) | 48.11% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | |
| 100.0% | 11.08% | 3.9e−05% | 0% | 0% | 0% | 0% | 0% | 0% | 0% | ||
| 100.0% | 100.0% | 88.78% | 4.613% | 0.001981% | 0% | 0% | 0% | 0% | 0% | ||
| 100.0% | 100.0% | 100.0% | 100.0% | 98.03% | 45.5% | 1.886% | 0.005933% | 2e−06% | 0% | ||
| 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 100.0% | 98.99% | 74.06% | 17.15% |
Figure 2: Comparing SNN to brute force search and tree-based methods.
Total index time (A, B) and average query time (C, D) for the synthetic uniformly distributed dataset, all in seconds, as the data size is varied (A, C) or the dimension is varied (B, D). Brute force query methods do not require an index construction, hence are omitted on the left. Our SNN method is the best performer in all cases, in some cases 10 times faster than the best tree-based method (balltree).For the second test we fix the number of data points at and vary the dimension . We perform queries for five selected radii as shown in Table 1. The table confirms that we have a wide variation in the number of returned data points relative to the overall number of points , ranging from empty returns to returning all data points. The indexing and query timings are shown in Fig. 2 (right). Again, among -d tree, balltree, and SNN, our method has the shortest indexing phase. The query time is obtained as an average over all query points and over all considered radii R. SNN performs best, with the average query time being between 3.5 and 6 times faster than balltree (the fastest tree-based method).
Comparison with GriSPy
GriSPy (Chalela et al., 2021), perhaps the most recent work on fixed-radius NN search, is an exact search algorithm which claims to be superior over the tree-based algorithms in SciPy. GriSPy indexes the data points into regular grids and creates a hash table in which the keys are the cell coordinates, and the values are lists containing the indices of the points within the corresponding cell. As there is an open-source implementation available, we can easily compare GriSPy against SNN. However, GriSPy has a rather high memory demand which forced us to perform a separate experiments with reduced data sizes and dimensions as compared to the ones in the previous “Near Neighbor Query on Synthetic Data”.
Again we consider uniformly distributed data points in , but now with (i) varying data size from to and averaging the runtime of five different radius queries with , and (ii) varying dimension over . The precise parameters and the corresponding ratio of returned data points are listed in Table 2. All queries are repeated 1,000 times and timings are averaged. Both experiments (i) and (ii) use the same query size as the index size.
| 1,000 | 2,154 | 4,641 | 10,000 | 21,544 | 46,415 | 100,000 | ||
|---|---|---|---|---|---|---|---|---|
| Varying ( ) | 0.05% | 0.05% | 0.048% | 0.049% | 0.05% | 0.05% | 0.049% | |
| 0.37% | 0.37% | 0.36% | 0.38% | 0.37% | 0.37% | 0.37% | ||
| 1.2% | 1.2% | 1.2% | 1.2% | 1.2% | 1.2% | 1.2% | ||
| 2.6% | 2.6% | 2.6% | 2.7% | 2.7% | 2.6% | 2.6% | ||
| 4.8% | 4.8% | 4.8% | 4.9% | 4.9% | 4.9% | 4.7% | ||
| 2 | 3 | 4 | ||||||
| Varying ( 10,000) | 0.75% | 0.05% | 0.0029% | |||||
| 2.9% | 0.38% | 0.042% | ||||||
| 6.1% | 1.2% | 0.2% | ||||||
| 10% | 2.7% | 0.59% | ||||||
| 15% | 4.9% | 1.3% |
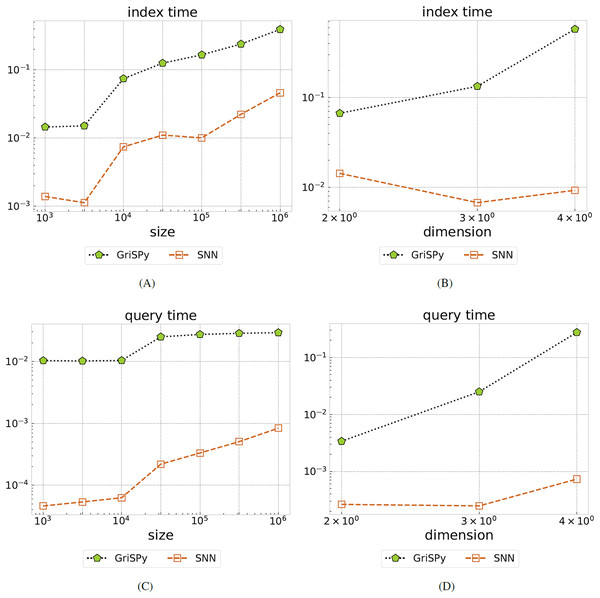
The index and query timings are illustrated in Fig. 3. We find that SNN indexing is about an order of magnitude faster than GriSPy over all tested parameters. For the experiment (i) where the data size is varied, we find that SNN is up to two orders of magnitude faster than GriSPy. For experiment (ii), we see that the SNN query time is more stable than GriSPy with respect to increasing data dimension.
Figure 3: Comparing GriSPy and SNN.
Total index time (A, B) and average query time (C, D) for on uniformly distributed data, all in seconds, as the data size is varied (A, C) or the dimension is varied (B, D). Our SNN method significantly outperforms GriSPy both in terms of indexing and query runtime.Near neighbor query on real-world data
We now compare various fixed-radius NN search methods on datasets from the benchmark collection by Aumüller, Bernhardsson & Faithfull (2020): Fashion-MNIST (abbreviated as F-MNIST), SIFT, GIST, GloVe100, and DEEP1B. Each dataset has an index set of points and a separate out-of-sample query set with points. See Table 3 for a summary of the data.
| Dataset | Dimension | Distance | Index size | Query size | Related reference |
|---|---|---|---|---|---|
| F-MNIST | 784 | Euclidean | 25,000 | 10,000 | Xiao, Rasul & Vollgraf (2017) |
| SIFT10K | 128 | Euclidean | 25,000 | 100 | Lowe (2004) |
| SIFT1M | 128 | Euclidean | 100,000 | 10,000 | Lowe (2004) |
| GIST | 960 | Euclidean | 1,000,000 | 1,000 | Oliva & Torralba (2004) |
| GloVe100 | 100 | Angular | 1,183,514 | 10,000 | Pennington, Socher & Manning (2014) |
| DEEP1B | 96 | Angular | 9,990,000 | 10,000 | Yandex & Lempitsky (2016) |
Table 4 lists the timings for the index construction of the tree-based methods and SNN. For all datasets, SNN is least 5.9 times faster than balltree (the fasted tree-based method). Significant speedups are gained in particular for large datasets: for the largest dataset DEEP1B, SNN creates its index more than 32 times faster than balltree.
| Dataset | -d tree | Balltree | SNN |
|---|---|---|---|
| F-MNIST | 9,035 | 7,882 | 1,335 |
| SIFT10K | 720.5 | 662.1 | 79.1 |
| SIFT1M | 3,292 | 2,921 | 179 |
| GIST | 319,400 | 297,900 | 29,140 |
| GloVe100 | 41,210 | 39,800 | 1,549 |
| DEEP1B | 446,000 | 464,100 | 14,730 |
Note:
Lower is better and the best values are highlighted in bold.
The query times averaged over all points from the query set are listed in Table 5. We have included tests over different radii R in order to obtain a good order-of-magnitude variation in the number of returned nearest neighbors relative to the index size , assessing the algorithms over a range of possible scenarios from small to large query returns. See the return ratios listed in Table 5. Again, in all cases, SNN consistently performs the fastest queries over all datasets and radii. SNN is between about 6 and 14 times faster than balltree (the fastest tree-based method). For the datasets GloVe100 and DEEP1B, SNN displays the lowest speedup of about 1.6 compared to our brute force 2 implementation, indicating that for these datasets the sorting-based exclusion criterion does not significantly prune the search space. (These are datasets for which the angular distance is used, i.e., all data points are projected onto the unit sphere.) For the other datasets, SNN achieves significant speedups between 2.6 and 5.6 compared to brute force 2, owing to effective search space pruning.
| Dataset | R | Brute force 1 | Brute force 2 | -d tree | Balltree | SNN | |
|---|---|---|---|---|---|---|---|
| F-MNIST | 800 | 0.01524% | 302.8 | 43.99 | 146.3 | 110.3 | 7.765 |
| 900 | 0.04008% | 244.4 | 43.96 | 152.2 | 110.7 | 8.602 | |
| 1,000 | 0.09283% | 218.5 | 44.1 | 157.2 | 111.2 | 9.413 | |
| 1,100 | 0.1960% | 217.3 | 44.28 | 160.5 | 111.5 | 10.21 | |
| 1,200 | 0.3818% | 216.2 | 44.32 | 163.3 | 110.8 | 11.18 | |
| SIFT10K | 210 | 0.02296% | 19.04 | 4.187 | 15.88 | 12.55 | 1.112 |
| 230 | 0.04892% | 21.56 | 4.153 | 18.58 | 13.75 | 1.170 | |
| 250 | 0.1147% | 21.24 | 4.546 | 18.35 | 13.15 | 1.458 | |
| 270 | 0.2718% | 22.97 | 4.279 | 19.91 | 14.73 | 1.128 | |
| 290 | 0.5958% | 22.06 | 4.276 | 19.86 | 15.33 | 1.093 | |
| SIFT1M | 210 | 0.02661% | 75.82 | 16.10 | 45.71 | 35.11 | 4.525 |
| 230 | 0.05671% | 78.24 | 16.15 | 46.86 | 38.37 | 4.557 | |
| 250 | 0.1231% | 86.03 | 16.29 | 50.30 | 40.75 | 4.598 | |
| 270 | 0.2663% | 80.13 | 16.17 | 54.13 | 42.76 | 4.660 | |
| 290 | 0.5608% | 69.77 | 16.17 | 58.80 | 44.55 | 4.727 | |
| GIST | 0.80 | 0.1430% | 3,955 | 862.2 | 3,144 | 2,160 | 281.5 |
| 0.85 | 0.1977% | 3,966 | 861.4 | 3,182 | 2,164 | 293.9 | |
| 0.90 | 0.2723% | 3,941 | 861.6 | 3,206 | 2,171 | 305.8 | |
| 0.95 | 0.3762% | 3,817 | 861.7 | 3,223 | 2,178 | 316.8 | |
| 1.00 | 0.5234% | 3,759 | 861.4 | 3,237 | 2,183 | 326.8 | |
| GloVe100 | 0.30 | 0.04506% | 516.9 | 127.3 | 671.5 | 567.5 | 78.38 |
| 0.31 | 0.07888% | 514.1 | 126.9 | 673.2 | 561.8 | 79.47 | |
| 0.32 | 0.1438% | 514.7 | 126.8 | 670.6 | 564.9 | 76.83 | |
| 0.33 | 0.2755% | 520.1 | 126.5 | 674.9 | 561.0 | 77.27 | |
| 0.34 | 0.5507% | 522.0 | 127.8 | 674.6 | 562.2 | 77.00 | |
| DEEP1B | 0.22 | 0.04495% | 4,281 | 1,079 | 5,711 | 4,731 | 803.0 |
| 0.24 | 0.09332% | 4,229 | 1,065 | 5,677 | 4,704 | 704.8 | |
| 0.26 | 0.1891% | 4,202 | 1,082 | 5,732 | 4,683 | 719.9 | |
| 0.28 | 0.3761% | 4,230 | 1,080 | 5,765 | 4,755 | 734.3 | |
| 0.30 | 0.7341% | 4,274 | 1,084 | 5,644 | 4,810 | 723.1 |
Note:
The search radius is R and is the average ratio of returned data points relative to the overall number of data points . Lower is better and the best values are highlighted in bold.
An application to clustering
We now wish to demonstrate the performance gains that can be obtained with SNN using the DBSCAN clustering method (Campello et al., 2015; Jang & Jiang, 2019) as an example. To this end we replace the nearest neighbor search method in scikit-learn’s DBSCAN implementation with SNN. To enure all variants perform the exact same NN queries, we rewrite all batch NN queries into loops of single queries and force all computations to run in a single threat. Except these modifications, DBSCAN remains unchanged and in all case returns exactly the same result when called on the same data and with the same hyperparameters (eps and min_sample).
We select datasets from the UCI Machine Learning Repository (Dua & Graff, 2017); see Table 6. All datasets are pre-processed by z-score standardization (i.e., we shift each feature to zero mean and scale it to unit variance). We cluster the data for various choices of DBSCAN’s eps parameter and list the measured total runtime in Table 7. The parameter eps has the same interpretation as SNN’s radius parameter R. In all cases, we have fixed DBSCAN’s second hyperparameter min_sample at . The normalized mutual information (NMI) (Cover & Thomas, 2006) of the obtained clusterings is also listed in Table 7.
| Dataset | Size | Dimension | #Labels | Related references |
|---|---|---|---|---|
| Banknote | 1,372 | 4 | 2 | Dua & Graff (2017) |
| Dermatology | 366 | 34 | 6 | Dua & Graff (2017), Güvenir, Demiröz & Ilter (1998) |
| Ecoli | 336 | 7 | 8 | Dua & Graff (2017), Nakai & Kanehisa (1991, 1992) |
| Phoneme | 4,509 | 256 | 5 | Hastie, Tibshirani & Friedman (2009) |
| Wine | 178 | 13 | 3 | Forina et al. (1998) |
| Dataset | eps | NMI | Brute force | -d tree | Balltree | SNN |
|---|---|---|---|---|---|---|
| Banknote | 0.1 | 0.05326 | 1,914 | 463.9 | 431.9 | 30.20 |
| 0.2 | 0.2198 | 1,739 | 454.5 | 434.3 | 48.67 | |
| 0.3 | 0.3372 | 1,968 | 452.0 | 438.8 | 50.61 | |
| 0.4 | 0.5510 | 1,759 | 457.5 | 442.4 | 51.21 | |
| 0.5 | 0.08732 | 1,752 | 477.2 | 449.8 | 53.01 | |
| Dermatology | 5.0 | 0.5568 | 706.8 | 138.8 | 124.3 | 86.81 |
| 5.1 | 0.5714 | 654.0 | 142.2 | 127.8 | 64.62 | |
| 5.2 | 0.5733 | 651.8 | 139.2 | 123.2 | 63.74 | |
| 5.3 | 0.5796 | 650.7 | 138.2 | 121.5 | 60.75 | |
| 5.4 | 0.4495 | 638.6 | 138.4 | 121.2 | 58.17 | |
| Ecoli | 0.5 | 0.1251 | 506.9 | 116.0 | 104.9 | 7.674 |
| 0.6 | 0.2820 | 491.9 | 116.0 | 105.2 | 8.105 | |
| 0.7 | 0.3609 | 496.0 | 116.5 | 107.2 | 9.263 | |
| 0.8 | 0.4374 | 500.9 | 116.7 | 105.1 | 10.97 | |
| 0.9 | 0.1563 | 499.8 | 116.3 | 105.0 | 11.39 | |
| Phoneme | 8.5 | 0.5142 | 3,497 | 17,290 | 7,685 | 926.9 |
| 8.6 | 0.5516 | 3,511 | 17,480 | 7,738 | 954.1 | |
| 8.7 | 0.5836 | 3,300 | 17,490 | 7,727 | 937.9 | |
| 8.8 | 0.6028 | 3,257 | 17,600 | 7,768 | 975.2 | |
| 8.9 | 0.5011 | 3,499 | 17,570 | 7,734 | 1,065 | |
| Wine | 2.2 | 0.4191 | 73.37 | 64.02 | 56.70 | 5.753 |
| 2.3 | 0.4764 | 64.65 | 63.84 | 56.29 | 5.703 | |
| 2.4 | 0.5271 | 66.91 | 63.26 | 55.74 | 5.612 | |
| 2.5 | 0.08443 | 67.29 | 63.11 | 56.34 | 6.106 | |
| 2.6 | 0.07886 | 67.12 | 63.45 | 56.25 | 6.094 |
Note:
The DBSCAN radius parameter is eps and the achieved normalized mutual information is NMI. Best runtimes are highlighted in bold.
The runtimes in Table 7 show that DBSCAN with SNN is a very promising combination, consistently outperforming the other combinations. When compared to using non-batched and non-parallelized DBSCAN with balltree, DBSCAN with SNN performs between 3.5 and 16 times faster while returning precisely the same clustering results.
Conclusions
We presented a fixed-radius nearest neighbor (NN) search method called SNN. Compared to other exact NN search methods based on -d tree or balltree data structures, SNN is trivial to implement and exhibits faster index and query time. We also demonstrated that SNN outperforms different implementations of brute force search. Just like brute force search, SNN requires no parameter tuning and is straightforward to use. We believe that SNN could become a valuable tool in applications such as the MultiDark Simulation (Klypin et al., 2016) or the Millennium Simulation (Boylan-Kolchin et al., 2009). We also demonstrated that SNN can lead to significant performance gains when used for nearest neighbor search within the DBSCAN clustering method.
While we have demonstrated SNN speedups in single-threaded computations on a CPU, we believe that the method’s reliance on high-level BLAS operations makes it suitable for parallel GPU computations. A careful CUDA implementation of SNN and extensive testing will be subject of future work.