Morphological and molecular analyses of parasitic barnacles (Crustacea: Cirripedia: Rhizocephala) in Korea: preliminary data for the taxonomy and host ranges of Korean species
- Published
- Accepted
- Received
- Academic Editor
- Khor Waiho
- Subject Areas
- Biodiversity, Marine Biology, Parasitology, Taxonomy, Zoology
- Keywords
- Morphology, Phylogenetic analysis, Taxonomy, Parasitic barnacles, Host range, Korean Rhizocephala
- Copyright
- © 2021 Jung et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2021. Morphological and molecular analyses of parasitic barnacles (Crustacea: Cirripedia: Rhizocephala) in Korea: preliminary data for the taxonomy and host ranges of Korean species. PeerJ 9:e12281 https://doi.org/10.7717/peerj.12281
Abstract
Morphological and molecular analyses of Korean rhizocephalan barnacle species were performed to examine their host ranges and taxonomy. Morphological examination and molecular analysis of mtDNA cox1, 16S, and nuclear 18S rRNA sequences revealed nine rhizocephalan species from three genera of the two families, Sacculinidae and Polyascidae. Phylogenetic analysis of molecular sequences revealed two new species candidates in the genus Parasacculina, and three Sacculina species (S. pilosella, S. pinnotherae, and S. imberbis) were transferred to the genus Parasacculina. Examination of host ranges revealed higher host specificity and lower infestation rates in Korean rhizocephalan species than rhizocephalans from other geographic regions. This is the first report of the taxonomy, species diversity, and host ranges of Korean parasitic rhizocephalan barnacles based on their morphological and molecular analyses. More information from extensive sampling of parasitic barnacles from a wide range of crustacean host species is necessary to fully understand their taxonomy, prevalence on decapod hosts, and phylogenetic relationships among major rhizocephalan taxa.
Introduction
The Rhizocephala comprises morphologically highly modified parasitic barnacles that use a wide range of crustaceans (mostly decapods) as their hosts, mostly parasites on decapods. Members of this group have complex life cycles, usually involving a series of pelagic larval stages followed by an endoparasitic interna stage and a reproductive externa stage where many organ systems (e.g., respiratory, digestive, sensory, and excretory systems) degenerate (Høeg, 1992; Øksnebjerg, 2000). In contrast to other crustacean species, they have a very simplified external structure (externa) and lack segmentation and appendages in the parasitic stage (Høeg & Lützen, 1995). Due to the simplified morphology of the externa, previous taxonomic studies of rhizocephalans have been based largely on larval morphology and the fine structure of externa observed from paraffin sectioning (Yoshida et al., 2011; Kobayashi et al., 2018), with further validation through DNA barcoding analysis (Yoshida et al., 2012; Yoshida, Hirose & Hirose, 2014; Høeg et al., 2019; Jung, Yoshida & Kim, 2019). Very recently, Høeg et al. (2019) modified the taxonomic system of Rhizocephala based on molecular phylogenetic analysis of 18S rDNA sequences, and additional molecular-based taxonomy of rhizocephalan barnacles was updated in Chan et al. (2021).
Since the first report of rhizocephalan barnacles (Krüger, 1912; Høeg & Lützen, 1985), northwestern Pacific species have been reported from southeast Russia (Korn et al., 2020), China (Li et al., 2015), Taiwan (Tu, Chan & Jeng, 2009; Yoshida et al., 2012), Japan (Shiino, 1943; Utinomi & Kikuchi, 1966; Nagasawa, Lützen & Kado, 1996), and Korea (Jung, Yoshida & Kim, 2019). Although Jung, Yoshida & Kim (2019) described 10 species of peltogastrid barnacles from 17 hermit crab species in Korea, the species diversity, distribution, taxonomy, and host range of other rhizocephalan parasitic barnacles are still largely unknown in Korea. In this study, we characterized nine rhizocephalan species, including two new cryptic species candidates, based on morphological examination and molecular analyses of mitochondrial (cytochrome c oxidase subunit I and 16S) and nuclear 18S rDNA sequences. In addition to the taxonomic accounts of the Korean species, we also investigated the prevalence of parasitic barnacles in decapod hosts and phylogenetic relationships among rhizocephalan species.
Methods
We examined the abdomens of 3,262 individuals of 25 Korean decapod host species collected from 16 sampling sites in Korea. In addition, 12 Japanese rhizocephalans from 11 sampling sites were obtained for comparative molecular study (Tables 1, 2). Korean voucher specimens in this study were deposited in the National Institute of Biological Resources (NIBR) and Honam the National Institute of Biological Resources (HNIBR). Japanese voucher specimens in this study were deposited in the Ryukyu University Museum, Fujukan, University of the Ryukyus, Okinawa, Japan (RUMF), and Coastal Beach of Natural History Museum and Institute, Chiba, Japan (CMNH).
| Host decapod species | Total number of individuals examined | Number of individuals infested | Infestation rate |
|---|---|---|---|
| Alpheus bisincisus | 16 | 0 | 0.0% |
| Arcotheres sinensis | 4 | 2 | 50.0% |
| Eualus sinensis | 16 | 0 | 0.0% |
| Gaetice depressus | 1400 | 14 | 1.0% |
| Helicana japonica | 28 | 0 | 0.0% |
| Hemigrapsus penicillatus | 50 | 0 | 0.0% |
| Hemigrapsus sanguineus | 114 | 9 | 7.9% |
| Hemigrapsus takanoi | 145 | 1 | 0.7% |
| Ilyoplax dentimerosa | 10 | 0 | 0.0% |
| Ilyoplax pusilla | 57 | 0 | 0.0% |
| Laomedia astacina | 15 | 0 | 0.0% |
| Macromedaeus distinguendus | 74 | 8 | 10.8% |
| Macrophthalmus (Mareotis) japonicus | 45 | 0 | 0.0% |
| Neotrypaea japonica | 35 | 0 | 0.0% |
| Pachygrapsus crassipes | 1 | 1 | 100.0% |
| Pagurus lanuginosus | 65 | 0 | 0.0% |
| Pagurus minutus | 811 | 0 | 0.0% |
| Pagurus nigrofascia | 28 | 0 | 0.0% |
| Palaemon serrifer | 17 | 0 | 0.0% |
| Parasesarma pictum | 61 | 0 | 0.0% |
| Pugettia intermedia | 14 | 1 | 7.1% |
| Scopimera globosa | 25 | 0 | 0.0% |
| Sestrostoma balssi | 16 | 0 | 0.0% |
| Stenalpheops anacanthus | 61 | 0 | 0.0% |
| Upogebia major | 154 | 2 | 1.3% |
| Total | 3262 | 38 | 1.2% |
All rhizocephalan specimens were fixed in 95% ethanol and subjected to morphological examination and molecular analysis. For morphological analysis, the externa and mantle were examined using an MZ8 dissection microscope (Leica, Wetzlar, Germany). Photographs were taken with a D200 digital camera (Nikon, Tokyo, Japan). Carapace length (cl) of the host decapod was measured as the length from the tip of the rostrum to the midpoint of the posterior margin of the carapace using a CD6CSX digital caliper (Mitutoyo, Kawasaki, Japan) to the nearest 0.1 mm.
For molecular analysis, the lateral end of the externa tissue of each rhizocephalan specimen was excised for total genomic DNA extraction using the QIAamp DNA Micro Kit (QIAGEN, Hilden, Germany). Universal primers LCO1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′) and HCO2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) were used to amplify a fragment of mitochondrial cytochrome c oxidase subunit I (cox1) (Folmer et al., 1994). To amplify the mitochondrial 16S rDNA gene, 16SH2 (5′-AGATAGAAACCAACCTGG-3′) and 16SL2 (5′-TGCCTGTTTATCAAAAACAT-3′) primers (Schubart, Neigel & Felder, 2000) were used. For PCR amplification of 18S rDNA, 18S-329R (TAATGATCCTTCCGCAGGTT) and 18S-AF (CAGCMGCCGCGGTAATWC) primers were used (Spears, Abele & Kim, 1992). Polymerase chain reaction (PCR) was performed in reaction volumes of 50 µL that included 2 µL DNA template, 5 µL 10 x Ex Taq Buffer, 2 µL of each primer (10 µM), 0.25 µL Go Taq DNA polymerase (Promega, Madison City, WI, USA), 2.5 µL dNTP mix (10 mM), and 35.75 µL distilled H2O. PCR amplification was performed using the following steps: 5 min denaturation at 94 °C followed by 35 cycles of 30 s at 94 °C, 1 min at 52 °C, 1 min at 72 °C, and a final extension of 7 min at 72 °C. PCR products were visualized on 1% agarose gels and sequenced with an ABI PRISM 3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA). Nucleotide sequences of the three gene fragments (mtDNA cox1, 16S, and nuclear 18S rDNA) were analyzed and edited using Geneious v. 9.1.8 (Kearse et al., 2012) and aligned using ClustalW in the MEGA10 program (Kumar et al., 2018). Nucleotide sequences were deposited in GenBank (mtDNA cox1: MZ216468–MZ216513; 16S: MZ215675–MZ215720; 18S rDNA: MZ215557–MZ215602). Forty-six additional rhizocephalan sequences of Sacculinidae and Polyascidae species available in GenBank were downloaded and included in the phylogenetic analyses (Table 2).
| Species | Host species | Location | Specimen number | cox1GenBankaccession no | 16S rDNA GenBank accession no | 18S rDNA GenBank accession no |
|---|---|---|---|---|---|---|
| Boschmaella japonica* | Chthamalus challengeri | Jôgashima, Japan | ZMUC CRU-3877 | AY265369 | ||
| Briarosaccus regalis* | Paralithodes camtschaticus | Alaska, USA | KR812178 | KR812157 | ||
| Clistosaccus paguri* | Germany | KT208500 | ||||
| Heterosaccus californicus* | Loxorhynchus grandis | CA, USA | AY520756 | |||
| Santa Barbara, USA | ZMUC CRU-3875 | AY265359 | ||||
| Heterosaccus dollfusi* | Charybdis longicollis | Mediterranean | FJ481949 | |||
| Portunus pelagicus | Sri Lanka | KY030832 | ||||
| Heterosaccus lunatus* | Charybdis callianassa | Moreton Bay, Australia | DQ059778 | FJ481947 | ||
| Heterosaccus papillosus* | Charybdis bimaculata | Korea | FJ481948 | |||
| Loxothylacus panopaei* | Rhithropanopeus harrisii | Neuse River, USA | ZMUC CRU-3876 | FJ481956 | AY265364 | |
| xanthoid crabs | USA | HQ848065 | ||||
| Loxothylacus texanus* | xanthoid crabs | USA | HQ848066 | |||
| Mycetomorpha vancouverensis* | Alaska, USA | MH974513 | ||||
| Parasacculina beauforti* | Scylla olivacea | Malaysia | KX426583 | |||
| Parasacculina compressa* | Ozius tuberculosus | Panglao, Philippines | KF561276 | |||
| Parasacculina granifera* | Portunus pelagicus | Moreton Bay, Australia | DQ059779 | |||
| Parasacculina imberbis comb. nov. | Pachygrapsus crassipes | Namhae, Korea | Korea 4 | MZ216470 | MZ215675 | MZ215557 |
| Shirosaki, Japan* | S2 | AB197804 | ||||
| Parasacculina leptodiae* | Leptodius affinis | Labrador, Singapore | ZMUC CRU-3870 | FJ481952 | AY265365 | |
| Parasacculina oblonga* | Cyclograpsus intermedius | Amakusa, Japan | G4028 | DQ059780 | ||
| Tomioka, Japan | ZMUC CRU-3871 | FJ481953 | AY265367 | |||
| Parasacculina pilosella comb. nov. | Pugettia intermedia | Sacheon, Korea | VSJAIV0000000010 (Korea 3) | MZ215679 | MZ215561 | |
| Parasacculina pinnotherae comb. nov. | Arcotheres sinensis | Imabari, Japan | CMNH-ZC-02762 (Japan 9) | MZ216499 | MZ215676 | MZ215558 |
| Busan, Korea | Korea 1 | MZ216468 | MZ215677 | MZ215559 | ||
| Korea 2 | MZ216469 | MZ215678 | MZ215560 | |||
| Parasacculina sinensis* | Leptodius affinis | Hong Kong, China | ZMUC CRU-3874 | AY265360 | ||
| Parasacculina shiinoi | Upogebia major | Namhae, Korea | Korea 25 | MZ216486 | MZ215562 | |
| Korea 26 | MZ216487 | MZ215680 | MZ215563 | |||
| Japan* | KF539761 | |||||
| Parasacculina sp. 1 | Macromedaeus distinguendus | Sacheon, Korea | Korea 15 | MZ216480 | MZ215683 | MZ215566 |
| Korea 16 | MZ216481 | MZ215684 | MZ215567 | |||
| Korea 17 | MZ216482 | MZ215685 | MZ215568 | |||
| Korea 20 | MZ216511 | MZ215686 | MZ215569 | |||
| Korea 21 | MZ216484 | MZ215687 | MZ215570 | |||
| Yeosu, Korea | Korea 14 | MZ216479 | MZ215682 | MZ215565 | ||
| Korea 19 | ||||||
| Parasacculina sp. 2 | Macromedaeus distinguendus | Sacheon, Korea | Korea 18 | MZ216483 | MZ215688 | MZ215571 |
| Parasacculina sp. | Guinusia dentipes | Tateyama, Japan | CMNH-ZC-02756 (Japan 6) | MZ216497 | MZ215681 | MZ215564 |
| Parasacculina yatsui | Hemigrapsus sanguineus | Taean, Korea | VSJAIV0000000011 (Korea 36) | MZ216507 | MZ215694 | MZ215576 |
| Toga Bay, Japan* | MG822656 | |||||
| Tongyeong, Korea | Korea 38 | MZ215695 | MZ215577 | |||
| Korea 39 | MZ216509 | MZ215696 | MZ215578 | |||
| Pachygrapsus crassipes | Misaki, Japan | CMNH-ZC-02770 (Japan13) | MZ216503 | MZ215691 | MZ215574 | |
| Tateyama, Japan | CMNH-ZC-02764 (Japan10) | MZ216500 | MZ215689 | MZ215572 | ||
| Katsuura, Japan | CMNH-ZC-02758 (Japan 7) | MZ215692 | ||||
| Tateyama, Japan | CMNH-ZC-02760 (Japan 8) | MZ216498 | MZ215693 | MZ215575 | ||
| Shirosaki, Japan* | S13 | AB197810 | ||||
| Tateyama, Japan | CMNH-ZC-02768 (Japan 12) | MZ216502 | MZ215690 | MZ215573 | ||
| Peltogasterella gracilis* | Pagurus filholi | Gyeongju, Korea | MADBK 160707_039 | MK604152 | ||
| Pagurus pectinatus | Busan, Korea | MADBK 430103_002 | MK604172 | |||
| Peltogaster postica* | Pagurus angustus | Chisi, Taiwan | NMNS-6795-003 | AB778096 | ||
| Pagurus filholi | Jeju, Korea | MADBK 430102_002 | MK604144 | |||
| Peltogaster reticulata* | Pagurus minutus | Namhae, Korea | MADBK 160706_065 | MK604167 | ||
| Vostok Bay, Russia | MN193579 | |||||
| Polyascus gregarius* | Eriocheir japonica | Maruyama, Japan | ZMUC CRU-3869 | AY265363 | ||
| Polyascus cf. gregarius | Hemigrapsus sanguineus | Namhae, Korea | VSJAIV0000000013 (Korea 27) | MZ216488 | MZ215697 | MZ215579 |
| Korea 30 | MZ216513 | MZ215700 | MZ215582 | |||
| Sacheon, Korea | Korea 29 | MZ216490 | MZ215699 | MZ215581 | ||
| Taean, Korea | VSJAIV0000000012 (Korea 37) | MZ216508 | MZ215703 | |||
| Yeosu, Korea | VSJAIV0000000014 (Korea 28) | MZ216489 | MZ215698 | MZ215580 | ||
| Korea 31 | MZ216491 | MZ215701 | MZ215583 | |||
| Hemigrapsus takanoi | Namhae, Korea | Korea 32 | MZ216492 | MZ215702 | MZ215584 | |
| Polyascus planus | Grapsus albolineatus | Nakagusuku, Okinawa, Japan | RUMF-ZC-7303 (Japan 2) | MZ216494 | MZ215705 | MZ215586 |
| Kenting, Taiwan* | ZMUC CRU-3872 | FJ481954 | AY265368 | |||
| Metopograpsus messor | Nago, Okinawa, Japan | RUMF-ZC-7305 (Japan 3) | MZ216495 | MZ215706 | MZ215587 | |
| Iriomote, Japan | RUMF-ZC-7309 (Japan 5) | MZ216496 | MZ215707 | MZ215588 | ||
| Nago, Okinawa, Japan | RUMF-ZC-7301 (Japan 1) | MZ216493 | MZ215704 | MZ215585 | ||
| Polyascus polygeneus* | Hemigrapsus sanguineus | Ôyano, Japan | ZMUC CRU-3873 | AY265362 | ||
| Sacculina carcini* | Carcinus maenas | Gullmar Fjord, Sweden | ZMUC CRU-3867 | FJ481957 | AY265366 | |
| Sacculina confragosa | Gaetice depressus | Namhae, Korea | Korea 11 | MZ216477 | MZ215709 | MZ215590 |
| VSJAIV0000000015 (Korea 12) | MZ216478 | MZ215710 | MZ215591 | |||
| Sacheon, Korea | Korea 5 | MZ216471 | MZ215715 | MZ215597 | ||
| Korea 6 | MZ216472 | MZ215716 | MZ215598 | |||
| Korea 7 | MZ216473 | MZ215717 | MZ215599 | |||
| Korea 8 | MZ216474 | MZ215718 | MZ215600 | |||
| Korea 9 | MZ216475 | MZ215719 | MZ215601 | |||
| Korea 10 | MZ216476 | MZ215708 | MZ215589 | |||
| Korea 23 | MZ216485 | MZ215712 | MZ215593 | |||
| Korea 24 | MZ216512 | MZ215594 | ||||
| Tongyeong, Korea | VSJAIV0000000016 (Korea 33) | MZ216504 | MZ215713 | MZ215595 | ||
| Korea 34 | MZ216505 | MZ215714 | MZ215596 | |||
| Korea 35 | MZ216506 | |||||
| Yeosu, Korea | Korea 13 | MZ216510 | MZ215711 | MZ215592 | ||
| Pachygrapsus crassipes* | Shirama, Japan | ZMUC CRU-3868 | AY265361 | |||
| Shirosaki, Japan | S22 | AB197803 | ||||
| Sacculina insueta* | Ptychognathus riedelii | Kawasan, Philippines | KF561274 | |||
| Sacculina upogebiae* | KF539762 | |||||
| Sacculinidae sp. | Thalamita sp. | Tateyama, Japan | CMNH-ZC-02766 (Japan 11) | MZ216501 | MZ215720 | MZ215602 |
| Sesarmaxenos gedehensis* | Sesarmops sp. | Kawasan, Philippines | KF561270 | |||
| Sylon hippolytes* | MG313989 |
Notes:
Phylogenetic relationships among rhizocephalan species were inferred for each of the three genes using maximum likelihood (ML) analysis and Bayesian inference (BI) implemented in RaxML version 8 (Stamatakis, 2014) and MrBayes v3.2.6 (Ronquist, Huelsenbeck & Teslenko, 2011), respectively. Phylogenetic trees were modified by MEGA 10. Maximum likelihood analyses of cox1, 16S, and 18S rDNA sequences were performed based on the Tamura-Nei (TN93) (Tamura & Nei, 1993), general time reversible (Tavaré, 1986), and Kimura 2-parameter (Kimura, 1980) models, respectively, with a gamma distribution (+G) and invariable sites (+I) rate categories based on Bayesian Information Criterion (BIC) scores model using the Model Selection option of MEGA10. The robustness of individual nodes in the ML trees was assessed by analysis of 1,000 bootstrap replications. Interspecific and intraspecific sequence divergences were estimated based on the K2P distance matrix in MEGA10.
Results
Based on morphological examination (shape and number of externae and mantle aperture) and mitochondrial sequence information, we identified 38 rhizocephalan individuals belonging to nine species, three genera, and two families isolated from eight decapod hosts species collected from 16 sites (Fig. 1; Table 2). All rhizocephalans identified by this study except Parasacculina pinnotherae comb. nov. were first reported from Korea. Detailed information regarding the Korean rhizocephalan species and their externa morphology is provided in Table 3.
Taxonomic accounts and morphological features of Korean rhizocephalan species
| Sacculinidae Lilljeborg, 1861 |
| SacculinaThompson, 1836 |
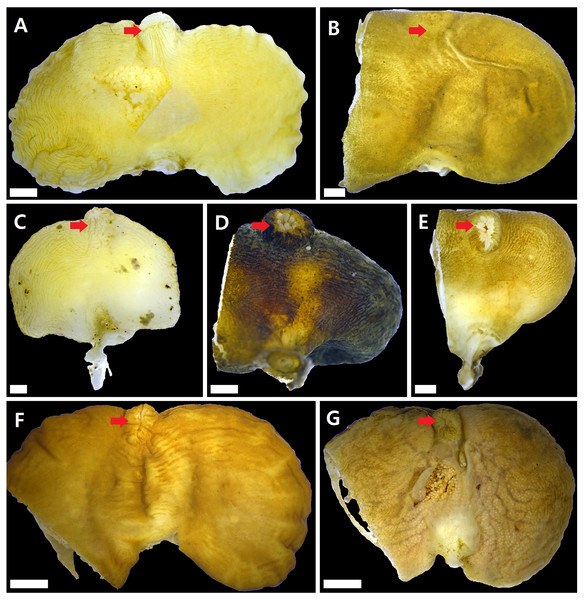
Sacculina confragosa Boschma1933 (Fig. 2A)
Materials examined: on Gaetice depressus: 1 ind., Sacheon (34.9 N 128.1 E), Korea 5, host: ♀, cl 11.5 mm; 1 ind. (2 externa), Sacheon (34.9 N 128.1 E), Korea 6, host: ♀, cl 11.8 mm; 1 ind., Sacheon (34.9 N 128.0 E), Korea 7, host: ♀, cl 13.7 mm; 1 ind., Sacheon (34.9 N 128.0 E), Korea 8, host: ♀, cl 9.1 mm; 1 ind., Sacheon (34.9 N 128.1 E), Korea 9, host: ♀, cl 10.3 mm; 1 ind., Sacheon (34.9 N 128.1 E), Korea 10, host: ♀, cl 11.8 mm; 1 ind., Namhae (34.7 N 127.9 E), Korea 11, host: ♂, cl 11.9 mm, feminization; 1 ind. (2 externa), Namhae (34.7 N 127.9 E), VSJAIV0000000015, Korea 12, host: ♀, cl 7.7 mm; 1 ind., Yeosu (34.7 N 127.8 E), Korea 13, host: ♂, cl 11.2 mm, feminization; 1 ind., Sacheon (34.9 N 128.0 E), Korea 23, host: ♂, cl 12.0 mm; 1 ind., Sacheon (34.9 N 128.0 E), Korea 24, host: ♂, cl 13.9 mm; 1 ind., Tongyeong (34.8 N 128.4 E), VSJAIV0000000016, Korea 33, host: ♀, cl 7.1 mm; 1 ind., Tongyeong (34.8 N 128.4 E), Korea 34, host: ♂, cl 14.9 mm; 1 ind., Tongyeong (34.6 N 128.5 E), Korea 35, host: ♂, cl 13.5 mm.
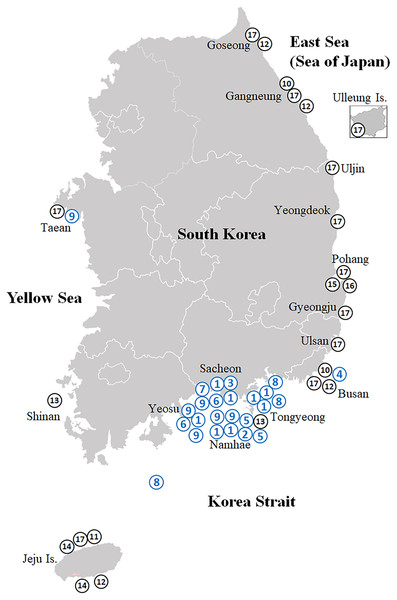
Figure 1: Map showing the collection sites of the Korean rhizocephalan species.
Numbered circles indicate sampling localities where rhizocephalan species were sampled in this study (blue) and Jung, Yoshida & Kim (2019); black). 1, Sacculina confragosa; 2, Parasacculina imberbis; 3, Parasacculina pilosella; 4, Parasacculina pinnotherae; 5, Parasacculina shiinoi; 6, Parasacculina sp. 1; 7, Parasacculina sp. 2; 8, Parasacculina yatsui; 9, Polyascus cf. gregarius; 10, Peltogaster lineata; 11, Peltogaster postica; 12, Peltogaster aff. ovalis; 13, Peltogaster aff. reticulatus; 14, Peltogaster sp. 1; 15, Peltogaster sp. 2; 16, Peltogaster sp. 3; 17, Peltogasterella gracilis.| Species | Externa | Mantle aperture | ||
|---|---|---|---|---|
| Shape | Externa number | Projection | Opening | |
| Sacculina confragosa | Wrinkled flat cordiform | Single or double | Elevated tube | Circular |
| Parasacculina imberbis comb. nov. | Smooth round-rectangular | Single | ||
| Parasacculina pilosella comb. nov. | Smooth and slightly flat oval | Single | Flat | Circular |
| Parasacculina pinnotherae comb. nov. | Smooth or slightly wrinkled flat oval or cordiform | Single or double | Elevated | Dot shaped |
| Parasacculina shiinoi | Smooth oval | Single | ||
| Parasacculina sp. 1 | Smooth or slightly wrinkled oval | Single | Elevated | Circular |
| Parasacculina sp. 2 | Smooth oval | Single | Slightly elevated | Circular |
| Parasacculina yatsui | Smooth or slightly wrinkled flat oval or flat cordiform | Single | Elevated | Slit shaped |
| Polyascus cf. gregarius | Smooth or slightly wrinkled flat cordiform | Single | Elevated | Slit shaped |
Figure 2: Externae of Korean rhizocephalans.
Red arrow: mantle, scale bar: two mm. (A) Sacculina confragosa. (B) Parasacculina pilosella comb. nov. (C) Parasacculina pinnotherae comb. nov. (D) Parasacculina sp. 1. (E) Parasacculina sp. 2. (F) Parasacculina yatsui. (G) Polyascus cf. gregarius. Externae of some specimens (B, D–G) were incomplete, in case they were used for molecular analysis (B, D–G).Host species: G. depressus, Pachygrapsus crassipes (Grapsidae), Cyclograpsus intermedius (Varunidae).
Distribution: Japan, Korea.
Diagnosis of the externa: whole externa mostly single and occasionally double, wrinkled cordiform with flat half-oval-shaped left and right lobes divided by an outer mid-groove and inner mid-ridge; outermost part of the robe wrinkled. Mantle well elevated, tube-shaped, and vertically slightly wrinkled with a circular opening at the extremity.
Remarks: Morphological characteristics of the examined materials correspond with their original description (Boschma, 1933) except for the number of externa. Some of our specimens (Korea 6, Korea 12) had double externae (15% of total examined individuals), whereas others had a single externa. This type of variation in the number of externa has been reported in a previous study (Shiino, 1943). This species is found most abundantly parasitizing medium-sized individuals of host crab species. Further study is needed to determine if this species is a predominant parasitic form on medium-sized host individuals.
Parasacculina imberbis (Shiino, 1943) comb. nov.
Polyascidae Høeg & Glenner in Høeg et al., 2019
Parasacculina Høeg & Glenner in Høeg et al., 2019
Materials examined: 1 ind., Namhae (34.7 N 128.0 E), Korea 4, host: ♀, cl 12.5 mm.
Host species: Pachygrapsus crassipes (Grapsidae).
Distribution: Japan, Korea.
Diagnosis of the externa: whole externa smooth, single, with a rounded-rectangular shape.
Remarks: The examined specimen had a single externa, but we were not able to examine morphological characteristics in more detail due to the immature stage of the specimen. Species identification of this specimen and its taxonomic placement in the genus Parasacculina were based on molecular analyses of mtDNA cox 1, 16S, and nuclear 18S gene sequences (Figs. 3A–3C; see Discussion for more details).
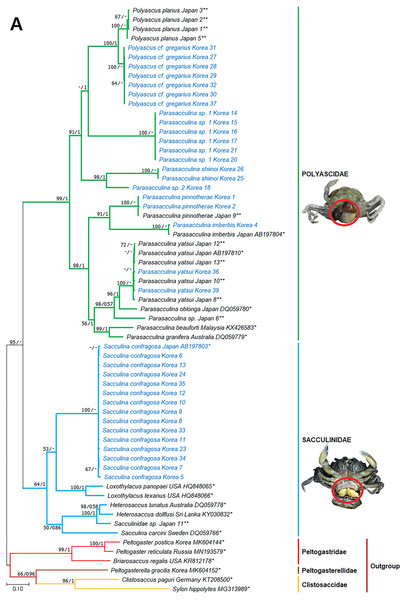
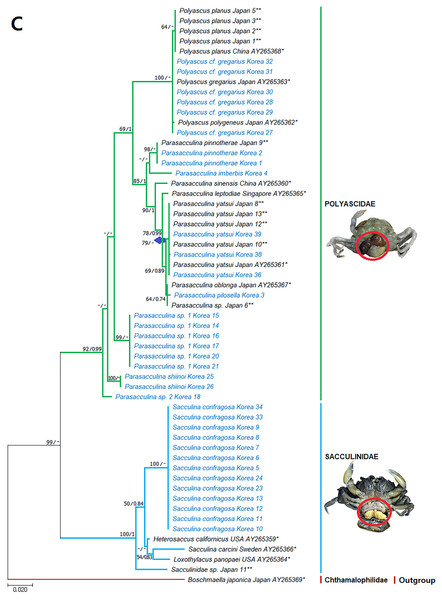
Figure 3: Phylogenetic tree of cox1 from rDNA from Korean rhizocephalan species using Maximum likelihood and Bayesian inference methods.
Values on nodes indicate maximum likelihood bootstrap support/Bayesian posterior probability. Sequences from Korean species determined in this study are indicated in blue. *: sequences derived from GenBank, **: Japanese sequences obtained in this study.Figure 4: Phylogenetic tree of 16S rDNA from Korean rhizocephalan species using Maximum likelihood and Bayesian inference methods.
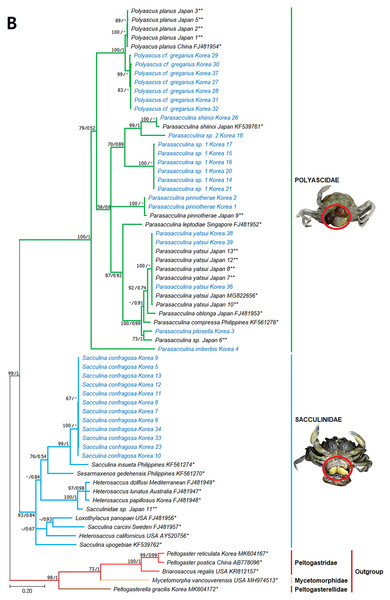
Values on nodes indicate maximum likelihood bootstrap support/Bayesian posterior probability. Sequences from Korean species determined in this study are indicated in blue. *: sequences derived from GenBank, **: Japanese sequences obtained in this study.Figure 5: Phylogenetic tree of 18S rDNA from Korean rhizocephalan species using Maximum likelihood and Bayesian inference methods.
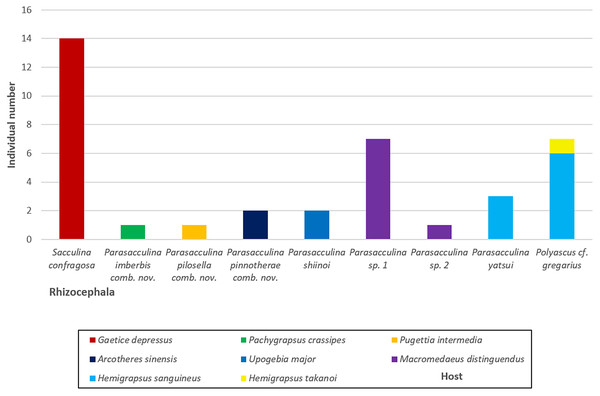
Values on nodes indicate maximum likelihood bootstrap support/Bayesian posterior probability. Sequences from Korean species determined in this study are indicated in blue. *: sequences derived from GenBank, **: Japanese sequences obtained in this study.Figure 6: A vertical bar chart showing the individual number of nine Korean rhizocephalan species found from their hosts.
Parasacculina pilosella (Kampen & Boschma, 1925) comb. nov. (Fig. 2B)
Material examined: 1 ind., Sacheon (34.9 N 128.1 E), VSJAIV0000000010, Korea 3, host: ♂, cl 13.7 mm.
Host species: Pugettia intermedia (Epialtidae).
Distribution: Indonesia, Japan, Korea.
Diagnosis of the externa: whole externa smooth, single, and slightly flat and oval. Mantle flat and vertically slightly wrinkled with a circular opening at the extremity.
Remarks: Four Sacculina species (S. muricata Boschma, 1931, S. pugettiae (Shiino, 1943), S. reinhardi (Boschma, 1955), and S. pilosella) were previously reported to parasitize Pugettia spp.. Morphological characteristics of the examined specimen correspond with the original description of S. pillosella (Van Kampen & Boschma, 1925). However, phylogenetic analysis of mtDNA 16S and 18S rDNA sequences showed that this species is nested within Parasacculina species (Figs. 3B and 3C), separated from Sacculina species (S. confragosa, S. upogebiae, and S. carcini). Therefore, we consider this species a member of the genus Parasacculina (see Discussion for more details).
Parasacculina pinnotherae (Shiino, 1943) comb. nov. (Fig. 2C)
Materials examined: 1 ind., Busan (35.2 N 129.2 E), Korea 1, host: ♀, cl 9.1 mm, in the mussel; 1 ind. (2 externa), Busan (35.2 N 129.2 E), Korea 2, host: ♂, cl 6.9 mm, in the mussel.
Host species: Arcotheres sinensis (Pinnotheridae).
Distribution: Japan, Korea.
Diagnosis of the externa: whole externa smooth or slightly wrinkled, single or double, and flat oval or cordiform in shape; each outer-posterior margin elevated into a conical shape. Mantle slightly elevated and vertically wrinkled with a small round opening at the extremity.
Remarks: Two Sacculina species (S. pertenuis (Boschma, 1933) and S. pinnotherae) have been reported to be parasitic on Pinnotheres spp.. Morphological characters of examined specimens correspond with the original description of S. pinnotherae (Shiino, 1943).
However, phylogenetic analysis of mtDNA cox 1, 16S, and nuclear 18S rDNA sequences placed this species within the genus Parasacculina (Figs. 3A–3C), not in the genus Sacculina. Therefore, we treated this species as a member of the genus Parasacculina (see Discussion for more details). The host crab (Arcotheres sinensis) is known to parasitize bivalves, so P. pinnotherae comb. nov. is a secondary parasite that is rare in the ocean (McDermott, 2009).
Parasacculina shiinoi (Lützen et al., 2016)
Materials examined: 1 ind., Namhae (34.9 N 127.9 E), Korea 25, host: cl 8.8 mm; 1 ind., Namhae (34.9 N 127.8 E), Korea 26, host: cl 11.1 mm.
Host species: Upogebia major (Upogebiidae).
Distribution: Japan, Korea.
Diagnosis of the externa: whole externa smooth, single, oval in shape.
Remarks: The examined specimens had a single externa, but detailed morphological characteristics could not be determined because of the immaturity of the specimens examined. Lützen et al. (2016) reported that Sacculina upogebiae parasitizes Upogebia species. Molecular analysis of mtDNA 16S rDNA sequences revealed that this species grouped with P. shiinoi (GenBank accession no: KF539761: Fig. 3B) with very high sequence identity (98.9%).
Parasacculina sp. 1 (Fig. 2D)
Materials examined: 1 ind., Yeosu (34.7 N 127.8 E), Korea 14, host: ♀, cl 7.6 mm; 1 ind., Sacheon (34.9 N 128.0 E), Korea 15, host: ♀, cl 8.0 mm; 1 ind., Sacheon (34.9 N 128.0 E), Korea 16, host: ♂, cl 9.7 mm; 1 ind., Sacheon (34.9 N 128.0 E), Korea 17, host: ♂, cl 9.9 mm; 1 ind., Yeosu (34.7 N 127.8 E), Korea 19, host: ♂, cl 14.6 mm; 1 ind., Sacheon (34.9 N 128.0 E), Korea 20, host: ♂, cl 14.1 mm; 1 ind., Sacheon (34.9 N 128.0 E), Korea 21, host: ♂, cl 17.7 mm.
Host species: Macromedaeus distinguendus (Xanthidae).
Distribution: Korea.
Diagnosis of the externa: whole externa single, smooth or slightly wrinkled, and oval in shape. Mantle large, elevated, and vertically wrinkled with circular opening at the extremity.
Remarks: Parasacculina leptodiae and P. sinensis have been reported to be parasites of Leptodius affinis, the most phylogenetically similar host species to M. distinguendus among the currently known hosts of Rhizocephala. However, the specimens examined in this study differ in morphology and molecular sequences from P. leptodiae and P. sinensis. This species has a single externa, whereas P. leptodiae has multiple externae. In addition, this species has a large, elevated mantle aperture, but P. leptodiae and P. sinensis have a flat mantle (Guérin-Ganivet, 1911; Boschma, 1933). Phylogenetic analysis clearly showed that the cox 1, 16S, and 18S rDNA sequences of this species are different from those of P. leptodiae and P. sinensis and all other Parasacculina species included in the analyses (Figs. 3A–3C). Therefore, we considered this species to be a new species candidate of the genus Parasacculina (see Discussion for more details).
Parasacculina sp. 2 (Fig. 2E)
Material examined: 1 ind., Sacheon (34.9 N 128.1 E), Korea 18, host: ♂, cl 13.5 mm.
Host species: Macromedaeus distinguendus (Xanthidae).
Distribution: Korea.
Diagnosis of the externa: whole externa smooth, single, and oval in shape. Mantle slightly elevated with circular opening at extremity.
Remarks: Previously, P. leptodiae and P. sinensis were known to be parasites of L. affinis, the most phylogeneically similar host species to M. distinguendus among the currently known hosts of Rhizocephala. However, the examined specimen of this species differed in morphology and molecular sequence from all Parasacculina species including Parasacculina sp. 1. This species has one externa compared to the multiple externae of P. leptodiae. In addition, this species has a slightly elevated mantle aperture, whereas P. leptodiae and P. sinensis have a flat mantle (Guérin-Ganivet, 1911; Boschma, 1933), and Parasacculina sp. 1 has a well-elevated mantle. Furthermore, phylogenetic analysis of mtDNA cox 1, 16S, and nuclear 18S rDNA sequences distinguished this species from other Parasacculina species with 18.1–30.6% sequence divergence in cox1, 14.0–27.5% sequence divergence in 16S rDNA, and 1.9−4.2% sequence divergence in 18S rDNA (Figs. 3A–3C). Therefore, we treated this species as a new species candidate of the genus Parasacculina (see Discussion for more details).
Parasacculina yatsui (Boschma, 1936) (Fig. 2F)
Materials examined: on Hemigrapsus sanguineus: 1 ind., Yeosu (34.1 N 127.3 E), VSJAIV0000000011, Korea 36, host: ♀, cl 22.3 mm; 1 ind., Tongyeong (34.8 N 128.4 E), Korea 38, host: ♂, cl 16.9 mm, feminization; 1 ind., Tongyeong (34.6 N 128.5 E), Korea 39, host: ♂, cl 14.2 mm, feminization.
Host species: Pachygrapsus crassipes (Grapsidae), H. sanguineus (Varunidae).
Distribution: Japan, Korea.
Diagnosis of the externa: whole externa smooth or slightly wrinkled, single, and cordiform with flat half-oval-shaped left and right lobes divided by outer mid-groove and inner mid-ridge; outermost part of the robe wrinkled. Mantle tube-shaped, elevated, and slightly wrinkled with slit opening at extremity.
Polyascus cf. gregarius (Okada & Miyashita, 1935) (Fig. 2G)
Polyascus Glenner, Lützen & Takahashi, 2003
Materials examined: on H. sanguineus: 1 ind., Namhae (34.7 N 127.9 E), VSJAIV0000000013, Korea 27, host: ♀, cl 18.4 mm; 1 ind., Yeosu (34.7 N 127.8 E), VSJAIV0000000014, Korea 28, host: ♂, cl 29.8 mm, feminization; 1 ind., Sacheon (34.9 N 128.1 E), Korea 29, host: ♀, cl 9.8 mm; 1 ind., Namhae (34.9 N 127.8 E), Korea 30, host: ♂, cl 16.7 mm, feminization; 1 ind., Yeosu (34.7 N 127.8 E), Korea 31, host: ♂, cl 8.9 mm, feminization; 1 ind., Taean (36.8 N 126.1 E), Korea 37, host: ♂, cl 14.1 mm, feminization.
on H. takanoi: 1 ind., Namhae (34.9 N 127.9 E), Korea 32, host: ♂, cl 9.5 mm.
Host species: Hemigrapsus sanguineus, H. takanoi, Eriocheir japonica (Varunidae).
Distribution: Japan, Korea.
Diagnosis of the externa: whole externa smooth or slightly wrinkled, single, and flat-cordiform shaped with flat half-oval-shaped left and right lobes divided by an outer mid-groove and inner mid-ridge; outermost part of the robe smooth or slightly wrinkled. Mantle tube-shaped, elevated, and vertically wrinkled with slit-shaped opening at the extremity.
Remarks: Morphological characteristics of the examined materials correspond with their original description (Okada & Miyashita, 1935) except for the number of externa and the host species. All specimens examined in this study had a single externa, whereas P. gregarius has multiple externae. In addition, the host species (H. sanguineus and H. takanoi) differ from the host species reported for P. gregarius, namely E. sinensis. Nevertheless, this species is likely P. gregarius because 18S rDNA sequences of these specimens were identical to the GenBank sequences of P. gregarius (Fig. 3C). In addition, individual variation in the number of externa of rhizocephalans has also been reported previously (Reinhard, 1942; Shiino, 1943; Høeg & Lützen, 1985).
In phylogenetic trees (Figs. 3A–3C), Polyascus cf. gregarious was clustered with P. planus that is commonly found in Japan and Taiwan. These two species are similar in having a flat-cordiform shaped externa, but different in some aspects of morphology and host species: the former has an elevated mantle and single externa, while the latter has an underdeveloped mantle and multiple externae (Boschma, 1933). In addition, the Varunidae crabs (H. sanguineus and H. takanoi) are used as P. cf. gregarious hosts, whereas the Grapsidae crabs (Grapsus albolineatus and Metopograpsus messor) are known as P. planus host (Tu, Chan & Jeng, 2009). Morphological and host range variation among rhizocephalan species has been reported by previous studies (Høeg & Lützen, 1985; Jung, Yoshida & Kim, 2019), and thus further studies with broader taxon sampling of P. gregarious and P. planus are needed to confirm an accurate species delimitation in their morphology and host range.
Phylogenetic relationships among rhizocephalan species
Since only Sacculinidae and Polyascidae species were found in this study, we focused on phylogenetic relationships among rhizocephalan species in these two families. Totals of 34 cox1 (555 bp), 33 16S rDNA (474 bp), and 35 18S rDNA (1002 bp) sequences were used for phylogenetic analysis, and the resulting ML and Bayesian trees were consistent with each other in that Sacculinidae and Polyascidae were monophyletic (Figs. 3A–3C). In all phylogenetic trees, the sequences of Korean rhizocephalans species nested and/or clustered with sequences of the same species retrieved from GenBank (Figs. 3A–3C).
Parasacculina sp. 1 and 2 were recognized as new species candidates because they did not show sister relationships with other Parasacculina species (Figs. 3A–3C). In the 16S and 18S DNA trees, they were placed at different positions and separated from P. leptodiae and P. sinensis, which share the host family and have similar morphological characteristics (Figs. 3B and 3C). Parasacculina sp. 1 and Parasacculina sp. 2 formed a group with P. shiinoi, but their 16S pairwise sequence divergences from P. shiinoi were substantial, ranging from 18.2–20.9% for Parasacculina sp. 1 and 14.4–14.8% for Parasacculina sp. 2. This group (Parasacculina sp. 1, Parasacculina sp. 2, and P. shiinoi) was separate from P. leptodiae in the 16S tree, whereas it was basal to the remaining Polyascidae species, including P. leptodiae and P. sinensis, in the 18S tree. In the cox1 tree, Parasacculina sp. 1 formed a well-defined sister group to Polyascus species (P. cf. gregarious and P. planus), whereas Parasacculina sp. 2 was grouped with the Korean isolate of P. shiinoi showing 18.1% sequence divergence (Fig. 3A). Interspecific sequence differences of the two new species candidates from other Polyascidae species were 18.1–32.1% for cox1, 14.0–28.8% for 16S rDNA, and 1.9−4.7% for 18S rDNA. In contrast to the high interspecific sequence divergences discovered, there were no individual variations in cox1, 16S, and 18S rDNA sequences among Parasacculina sp. 1 specimens.
The three Sacculina species (S. imberbis, S. pilosella, and S. pinnotherae) clustered with Parasacculina species (Figs. 3A–3C): S. imberbis grouped with S. pinnotherae, that is sister to other Parasacculina species based on analysis of cox1 (P. yatsui, P. granifera) and 18S rDNA (P. yatsui, P. sinensis, P. leptodiae) sequences (Figs. 3A, 3C). S. pilosella formed a sister group to P. compressa, P. oblonga, and P. yatsui in the 16S and 18S rDNA trees (Figs. 3B and 3C). Interspecific sequence differences between Sacculina species and Parasacculina species were remarkably large, ranging from 27.3–33.4% for cox1, 27.5–34.9% for 16S rDNA, and 9.2–10.1% for 18S rDNA. In contrast, intraspecific sequence divergences were very low with a maximum sequence difference of 1.0% for cox1 and 0.2% sequence difference for 16S rDNA sequences among S. confragosa individuals. All S. pinnotherae individuals had identical cox1, 16S, and 18S rDNA sequences (Figs. 3A–3C).
Discussion
In this study, we identified nine species of Korean rhizocephalans from eight host decapod species using morphological and molecular analyses. Close examination of host ranges revealed that Korean rhizocephalan species have a different host prevalence than reported for rhizocephalan species from other geographic regions. In Korea, rhizocephalans were firstly found from three decapod hosts, i.e., Hemigrapsus takanoi, Macromedaeus distinguendus, and Pugettia intermedia. We also found that most Korean rhizocephalans showed high host specificity, parasitizing only one host, except Polyascus cf. gregarius that was found on two crab species (Fig. 4). The notable differences in host range between geographic isolates (i.e., rhizocephalans from Korea and other geographic regions) might be due to geographical variation in host species diversity and abundance or insufficient information about the geographic origins of host crab species as proposed by Jung, Yoshida & Kim (2019). In addition, unlike Korean S. confragosa individuals that were all found on only one grapsid crab species, Gaetice depressus, the Japanese form is known to parasitize three crab species, G. depressus, Pachygrapsus crassipes, and Cyclograpsus intermedius. Furthermore, Japanese P. yatsui parasitizes not only G. depressus, but also P. crassipes (Tsuchida, Lützen & Nishida, 2006; Kobayashi et al., 2018), whereas the Korean form of P. yatsui was found only on Hemigrapsus sanguineus. We could not determine if other crab species including P. crassipes and C. intermedius are potential hosts of Korean S. confragosa and P. yatsui because of the limited pool of crab host species examined in this study. Extensive taxon sampling of decapod hosts and their parasitic barnacles is needed to obtain a complete understanding of the host ranges of rhizocephalan barnacles and the distribution and prevalence of host-parasite associations.
The decapod host infestation rate of Korean rhizocephalan barnacles was much lower than that reported for Japanese species. In Japan, 35 individuals representing three rhizocephalan species were found in 354 individuals of three crab species, corresponding to an infestation rate of 9.9% (Tsuchida, Lützen & Nishida, 2006). By contrast, the infestation rate of Korean rhizocephalans was substantially lower at 1.2% on average (Table 1). Species richness and extent of host usage by parasitic barnacles are tightly correlated to the availability of host species (species diversity and abundance; Kamiya et al., 2014). Differences in the extent of host usage by rhizocephalan barnacles between the two geographic regions are likely due to differences in host species diversity and abundance, as well as the sample size of examined materials (e.g., total numbers of individuals and host species). Since we examined the prevalence of rhizocephalans on all decapod hosts (a total of 3,262 host individuals inspected), our result is likely an accurate estimate of the infestation rate. On the other hand, this prevalence difference between Korea and Japan may be originated from salinity, season, host sex and size (Mouritsen et al., 2018) or biogeographical differences (Kim et al., 2020). In a previous study, the infestation rate of Korean hermit crabs by rhizocephalans was reported to be 0.9% (Jung, Yoshida & Kim, 2019), which is similar to the infestation rate observed in this study. The unexpectedly high infestation rates (>50%) of Pachygrapsus crassipes and Arcotheres sinensis are due to strong bias from the very small sample size (one to four individuals) examined. The marine ecosystems in different geographic regions display different assemblages of barnacles (Kim et al., 2020) and thus extensive sampling of parasitic barnacles from a wide range of decapod host species is necessary to better understand their prevalence, infection intensity, and host range specificity (Mouritsen et al., 2018).
In addition to their host ranges, morphological and molecular analyses in this study provided insights into the taxonomy of Korean rhizocephalan barnacle species. Phylogenetic trees recognized four monophyletic rhizocephalan families, i.e., Polyascidae, Sacculinidae, Peltogastridae, and Peltogasterellidae, consistent with previous molecular analysis (Høeg et al., 2019) and morphology-based classification. Polyascidae is characterized by multiple externa and reproduces asexually (Glenner, Lützen & Takahashi, 2003), whereas Sacculinidae is characterized by single externa and sexual reproduction. Peltogastridae and Peltogasterellidae species mainly parasitize hermit crabs, and Peltogastridae is distinguished from Peltogasterellidae by the presence of the chitinous shield on its middle part of externae (Høeg et al., 2019). Two new species candidates in the genus Parasacculina (Parasacculina sp. 1 and Parasacculina sp. 2) were recognized based on molecular phylogenetic analyses. These species were distinct from their congeneric species, P. leptodiae and P. sinensis, based on phylogenetic analyses of mtDNA (16S rDNA) and nuclear (18S rDNA) sequences (Figs. 3B–3C) even though they are morphologically indistinguishable and were found in the same host species. These two species are genetically distinct cryptic species. Furthermore, we transferred three Korean Sacculina species (i.e., Sacculina imberbis, S. pilosella, and S. pinnotherae) to the genus Parasacculina because they grouped with Parasacculina species in mtDNA cox1, 16S, and 18S rDNA phylogenetic trees (Figs. 3A–3C). This new taxonomic replacement is consistent with previous studies that transferred several Japanese and Chinese Sacculina species to Parasacculina based on molecular evidence (Tsuchida, Lützen & Nishida, 2006; Glenner et al., 2010; Høeg et al., 2019).
Comparison of the external cuticles of Korean species with previously published morphological data provided new insight into the taxonomic status of the families Sacculinidae and Polyascidae. Although Høeg et al. (2019) showed that Sacculinidae and Polyascidae are phylogenetically distinct, the original descriptions of Polyascidae (Høeg et al., 2019) did not specify morphological characters differentiating this family from Sacculinidae. For example, Høeg et al. (2019) noted that polyascids have a smooth or almost smooth external cuticle, but some polyascid species (P. pinnotherae comb. nov. and P. yatsui) in the present study had wrinkled cuticles (Figs. 2C, 2F). In addition, Høeg et al. (2019) mentioned that Polyascus species have multiple externae, but Polyascus cf. gregarius in this study had only a single externa (Fig. 2G). These results indicate that the morphological characteristics of external cuticles, previously considered to be taxonomically valid features, are highly variable and cannot be used as diagnostic characters. Future comparative analyses of morphological characters along with molecular sequences are necessary to confirm the taxonomic status of Sacculinidae and Polyascidae and the taxonomic replacement of the three Korean Sacculina species in the genus Parasacculina.
Conclusions
In conclusion, this is the first report of the taxonomy, species diversity, and host ranges of Korean parasitic rhizocephalan barnacles based on morphological and molecular analyses. We identified nine parasitic barnacle species, including two new species candidates in the genus Parasacculina, in Korea. In addition, we found higher host specificity and lower infestation rates for Korean rhizocephalan species than reported for rhizocephalan species from other geographic regions. Nevertheless, the results of this study are based on preliminary data derived from limited taxon sampling in a narrow geographic range in Korea. Additional data from extensive samplings of parasitic barnacles from a wide range of crustacean host species are necessary to better understand the taxonomy, prevalence, host usage, and phylogenetic relationships of rhizocephalan species.