Genome-wide identification and expression analysis of the cucumber PYL gene family
- Published
- Accepted
- Received
- Academic Editor
- Ivo Feussner
- Subject Areas
- Agricultural Science, Bioinformatics, Biotechnology, Molecular Biology, Plant Science
- Keywords
- Abscisic acid, Cucumber, PYL gene, Abiotic stress
- Copyright
- © 2022 Zhang et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2022. Genome-wide identification and expression analysis of the cucumber PYL gene family. PeerJ 10:e12786 https://doi.org/10.7717/peerj.12786
Abstract
Abscisic acid (ABA) is a very important hormone in plants. It regulates growth and development of plants and plays an important role in biotic and abiotic stresses. The Pyrabactin resistance 1-like (PYR/PYL) proteins play a central role in ABA signal transduction pathways. The working system of PYL genes in cucumber, an important economical vegetable (Cucumis sativus L.), has not been fully studied yet. Through bioinformatics, a total of 14 individual PYL genes were identified in Chinese long ‘9930’ cucumber. Fourteen PYL genes were distributed on six chromosomes of cucumber, and their encoded proteins predicted to be distributed in cytoplasm and nucleus. Based on the phylogenetic analysis, the PYL genes of cucumber, Arabidopsis, rice, apple, Brachypodium distachyon and soybeancould be classified into three groups. Genetic structures and conserved domains analysis revealed that CsPYL genes in the same group have similar exons and conserved domains. By predicting cis-elements in the promoters, we found that all CsPYL members contained hormone and stress-related elements. Additionally, the expression patterns of CsPYL genes were specific in tissues. Finally, we further examined the expression of 14 CsPYL genes under ABA, PEG, salt stress. The qRT-PCR results showed that most PYL gene expression levels were up-regulated. Furthermore, with different treatments about 3h, the relative expression of PYL8 was up-regulated and more than 20 times higher than 0h. It indicated that this gene may play an important role in abiotic stress.
Introduction
ABA is one of the hormones that regulates growth and development of plants, participating in plant response to biological and abiotic stresses (Lee & Luan, 2012). The study found that abscisic acid not only participates in plant growth, such as seed dormancy, cell division and elongation, stomatal movement, embryo development, but also in response to biotic and abiotic stresses (Bartels & Sunkar, 2005; Finkelstein, Gampala & Rock, 2002). PYLs belong to the START (Star-related lipid-transfer) superfamily of ligand-binding proteins, which contains hydrophobic ligand cavity and can bind directly to ABA (Grill & Himmelbach, 1998; Szostkiewicz et al., 2010). ABA signal transduction pathway mainly consists of three parts: ABA receptor PYR/PYL/PCAR, negative regulator factor protein phosphatase PP2C (Ma et al., 2009; Park et al., 2009) and positive regulator factor protein kinase SnRK2 (Kobayashi et al., 2005). Normally PP2C binds to SnRK2, SnRK2 serine/threonine residues are dephosphorylated, protein kinases are inactivated, and the ABA signaling pathway is silenced (Ma et al., 2009; Vlad et al., 2009). When stimulated by external signals, ABA content increased and closely combined with the internal hydrophobic cavity of PYR / PYL / RCAR protein. This binding is accomplished by the formation of ion pairs between a lysine residue side chain of the receptor protein in the cavity and the carboxyl group of the ABA. After ABA combined with the receptor protein, the conformation of the receptor protein in hydrophobic cavity is changed. It is mainly reflected in two conservative rings, “gate ring” and “latch ring” (Fujii et al., 2009). After ABA binding, a position of the proline residue on the “gate ring” was shifted and to close the cavity, and a serine residue on the ring was squeezed out of the cavity by ABA, then a histidine residue on the “latch ring” is transferred into the cavity and binds to the ABA through van der Waals force and hydrogen bond, finally the “latch ring” conformation changes locked the “gate ring”, leads to ABA fixed in the cavity, and provides a binding site for the PYL protein and the PP2C (Szostkiewicz et al., 2010). The activity of PP2C bound with PYL was inhibited, the SnRK2 is separated from the PP2C, and then the downstream transcription factors are phosphorylated and ABA signaling pathway is initiated.
ABA and its required proteins are found in algae, but they lack ABA receptor and do not respond to ABA. Terrestrial plants are considered to be monophyletic and gradually evolve into ABA activated receptors in continuous evolution (Sun et al., 2019). The PYR/PYL/RCAR family contains 14 genes in Arabidopsis (Dupeux et al., 2011). According to phylogenetic tree, ABA receptors have been divided into three subfamilies: I, II, and III (Ma et al., 2009). Subfamilies I and II receptors are monomeric, whereas subfamily III receptors are dimeric and exclusive to the more recently diverged angiosperms (Umezawa et al., 2010). Subfamily III receptors require ABA for dimer dissociation, which results in low basal receptor activity in the absence of ABA (Dupeux et al., 2011). Furthermore, orthologous genes in other crops have been reported, including 13 PYLs in rice, 13 PYLs in maize, 27 PYLs in cotton, 14 PYLs in tomato, and 8 PYLs in grape (Yadav et al., 2020; He et al., 2018; Chen et al., 2017; González-Guzmán et al., 2014; Boneh et al., 2012). Moreover, the functions of some PYL genes have been successfully verified. For example, the AtPYL6 and AtPYL13 in Arabidopsis have been shown to inhibit seed germination (Fuchs et al., 2014), AtPYR1, AtPYL1-5, AtPYL8 can promote ABA induce seed germination, stomatal closure and root growth (Gonzalez-Guzman et al., 2012; Zhao et al., 2014). In addition to the PYL genes associated with growth, AtPYL4, AtPYL9 can improve the drought resistance of Arabidopsis (Pizzio et al., 2013; Zhao et al., 2016). Overexpression of OsPYL5 and OsPYL10 in rice can enhance drought resistance, salt tolerance and cold resistance (Kim et al., 2014). Moreover, the ectopic expression of OsPYL3 in Arabidopsis can enhance the cold tolerance and drought tolerance of Arabidopsis (Lenka et al., 2018). Overexpression of ZmPYL8, ZmPYL9 and ZmPYL12 in maize can enhance the cold resistance of maize (He et al., 2018). Excessive expression of GhPYL9-11A in cotton enhances drought resistance of cotton, and overexpression of GhPYL9-11A in transgenic Arabidopsis is sensitive to ABA during seed germination and early seedling stage (Chengzhen et al., 2017). These results indicated that PYL genes play a significant role in plants tolerance under abiotic stress. However, the information of PYL gene family in cucumber has not been reported.
Cucumber (Cucumis sativus L, 2n = 2X = 14) has a small number of chromosomes and small genome (Luming et al., 2012). The first assembly of the genome sequence has been completed (Guo et al., 2009). Cucumber is an annual vine or climbing herb of Cucurbitaceae Cucumis, which is one of the important vegetable crops in China. After long-term cultivation and domestication, it is widely distributed all over the world. During the process of cucumber cultivation, it is vulnerable to various biotic and abiotic stresses, which affect quality and yield. Therefore, this study identified and analyzed the cucumber PYL gene family and used quantitative PCR (qRT- PCR) to determine the expression of 14 cucumber PYL genes under ABA, salt stress and drought stress, which provided a theoretical basis for the future study of cucumber stress resistance.
Materials and Methods
Plant materials and treatment
The germinated seeds (Cucumis sativus L, “L306” cultivar) were treated by 5% NaClO 10 min and deionized water washing 3–5 times. After that, the cucumber seeds were put into an artificial climate incubator to promote germination. The cultivation condition: relative humidity 80%, temperature 25 °C/18 °C (day/night), and the light intensity during the day is 250 µmol m−2 s−1. After 4 days of culture, cucumber seedlings were soaked in Yamazaki nutrient solution for hydroponics. Cucumber seedlings were treated when they grew to three true leaves. This experiment set up four treatments (T1: 50 µmol/L ABA, T2: 100 μmol/L ABA, T3: 200 mmol/L NaCl, T4: 10% PEG). The cucumber leaves were collected after 0 h, 3 h, 6 h, 12 h and 24 h. Finally, put fresh samples in liquid nitrogen, immediately and stored in −80 °C refrigerator.
Genome-wide identification of cucumber PYL gene family
The protein sequences of 14 PYL genes in Arabidopsis were downloaded from the Arabidopsis information resource (TAIR) website (https://www.arabidopsis.org/). And then, the BLASTp comparison was carried out in cucumber genome data (http://cucurbitgenomics.org/), the retrieval threshold was set as E-value < E−10 (Altschul et al., 1997). Scaffold assemblies of three cucumber lines (Chinese long ‘9930’, Gy14, and B10) are available so far. This draft genome was assembled using Chinese long inbred line ‘9930’ version 3 (Cavagnaro et al., 2010). The login number of duplicate genes in the comparison results was manually deleted. Then the gene domain after screening was searched in the Pfam database (http://pfam.xfam.org/search#tabview=tab1). A total of 15 CsPYL genes were obtained. The Polyketide_cyc2 domains (PF10604) were found in all CsPYLs. After naming according to the chromosome order, it was found that CsPYL1 had an unknown domain and a large molecular weight, so it was removed. Then downloading HMM files for HMM search comparison, a total of 14 PYL genes family members in cucumber were identified. Finally, through NCBI-CDD database (https://www.ncbi.nlm.nih.gov/cdd/) (Marchler-Bauer et al., 2015) identified whether the candidate sequences have PYR/PYL (RCAR) like domain (cd07821).
Analysis of chromosome location and collinearity analysis
The Chinese long ‘9930’ V3 version of gff3 file was downloaded from cucumber genome database, and the chromosome location and chromosome length information of 14 PYL genes were screened out and named CsPYLs according to their distribution on chromosomes. The MG2C (http://mg2c.iask.in/mg2c_v2.0/) was used to map the chromosomal location. Using Mcscanx search for homology, the protein-coding genes from cucumber genome was compared against itself and Arabidopsis genomes using BLASTp and retrieval threshold was set as E-value < E−5. Others were not modified by default parameters. Whole-genome BLASTP results were used to compute collinear blocks for all possible pairs of chromosomes and scaffolds (Wang et al. 2012a). Subsequently, TBtools was used to highlight the identified PYL collinear pairs and their collinear pairs with Arabidopsis (Chen et al., 2020).
Selective pressure analysis of cucumber PYL gene family
PAL2NAL (http://www.bork.embl.de/pal2nal/index.cgi?) were used to calculate the non-synonymous/synonymous (dN/ds) value of duplicate gene pairs (Goldman & Yang, 1994).
Sequence analysis and basic information of cucumber PYL gene family
The subcellular localization of PYL protein in cucumber was predicted by Wolf PSORT (https://wolfpsort.hgc.jp/) (Xiong et al., 2015). The number of amino acids, isoelectric point (PI), molecular weight (MW) and other physical and chemical information of CsPYL genes were identified on the ExPASy website (https://web.expasy.org/protparam/) (Artimo et al., 2012). Additionally, the secondary structure of cucumber PYL protein were predicted through the online web site (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html).
Construction of phylogenetic tree
The phylogenetic tree of PYL gene family of cucumber, A. thaliana, rice, soybean, grape, B. distachyon and apple were constructed by using MEGA 7 software with muscle methods to align multiple sequences (Kumar, Stecher & Tamura, 2016). The amino acid substitution model was (JTT+G) (Supplemental Information 3), and the bootstrap values were calculated for 1,000 replicates. Finally, a stable minimum neighbor tree was selected to represent their evolutionary relationship and beautify the constructed tree by ITOL website (Ivica & Peer, 2016) (http://itol.embl.de/).
Analysis of gene exon-intron structures and protein conserved motifs
Upload CDS and genome-wide sequences of 14 cucumber PYL genes on GSDS (GSDS v2.0) websites (http://gsds.gao-lab.org/) (Yang et al., 2015) for genetic structure analysis; The conserved motifs of 14 cucumber PYL genes were analyzed by MEME website (http://meme-suite.org/tools/meme) (Bailey et al., 2009). The maximum motif number was set to 10, and the remaining parameters were the default values. Functional annotations of these motifs were performed using HHpred (https://toolkit.tuebingen.mpg.de/tools/hhpred) (Zimmermann et al., 2018).
Analysis of cis-acting elements in CsPYL gene promoters
In order to predict the cis-acting elements in the promoter of CsPYL genes, TBtools was used to extract the genomic sequences of 14 cucumber PYL genes up to 2,000 bp upstream of the initiation codon (ATG). Then, the 2,000 bp sequences of 14 CsPYLs were submitted to the online website plantcare for prediction (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot, 2002).
Transcriptome analysis of CsPYL genes in different tissues
In order to study the gene-specific expression of CsPYL genes in different tissues of cucumber, the accession number PRJNA80169 was used from cucumber genome data (http://cucurbitgenomics.org/) to obtain cucumber RNA samples from different tissues and organs (tendril-base, tendril, root, leaf, stems, ovary-unfertilized, ovary-fertilized, ovary) RNA-Seq data (Li et al., 2011; Harris et al., 2008). Finally, the method of log2 was used for data conversion and Tbtools software was used to draw the expression heat map of CsPYL gene.
RNA extraction and real-time RT-PCR
Total RNA was isolated from collected samples using a Plant RNA Extraction kit (Tiangen, China). The complementary DNA was synthesized by using fastking cDNA dispersing RT supermaxs kit (Tiangen, China) with 2 µl RNA as template. The CDS sequences of CsPYL genes were input into the homepage of Shanghai biology company (Shanghai, China) for online primer design (Table S1), and then the primer sequences were synthesized. The SYBR Green kit (Tiangen, China) was used for fluorescence quantitative analysis. The volume of reaction system was 20 µL, which contained 2 µL cDNA solution, 10 µL2∗SuperReal PreMix Plus, 0.6 µL of 10 µM forward and reverse primers, 0.4 µL 50∗ROX Reference Dye and 6.4 µL of distilled deionized water. Then the qRT-PCR was performed with LightCycler® 480 II real-time fluorescence quantitative PCR instrument. The amplification program conditions were as follows: 95 °C for 15 min, and 40 cycles of 95 °C for 10 s and 60 °C for 30 s. Each sample was replicated 3 times. The relative expressions of CsPYL genes were calculated by 2−ΔΔCT method (Livak & Schmittgen, 2002), with CsActin as an internal control (Gan et al., 2013; Wan et al., 2010). Each sample repeat at least three times.
SPSS 20.0 was used for one-way ANOVA, and Duncan method was used to test the significance of the difference (P < 0.05) and data were all the average of 3 replicates. The Excel was used to complete the histogram of relative expressions.
Results
Genome-wide analysis and chromosome distribution of CsPYL gene family
We determined that 14 putative CsPYL were present in the cucumber genome through BLASTP by using 14 AtPYL sequences as references. From the analysis of their physical and chemical properties (Table 1), PYL2 has the largest number of amino acids and the largest molecular weight of protein. The other members of PYL protein have 162∼233 amino acids and protein molecular weight range 18.17 from 25.66 KDa. In addition, isoelectric points (PI) are from 4.91 to 8.28. Only CsPYL4 is basic protein, and the others are acidic proteins. The total average hydrophobic index of 14 CsPYL genes family members were less than zero, all of them were hydrophilic proteins.
| Gene ID | Gene name | Chromosome | Gene location | Amino acid number |
Molecular weight (kd) |
PI | Instability index | Aliphatic index | Grand average of hydropathicity | |
|---|---|---|---|---|---|---|---|---|---|---|
| Start position | End position | |||||||||
| CsaV3_2G030780.1 | PYL1 | 2 | 20234462 | 20235706 | 162 | 18.17 | 5.43 | 43.43 | 72.22 | −0.246 |
| CsaV3_2G030790.1 | PYL2 | 2 | 20234462 | 20239446 | 344 | 38.47 | 5.25 | 37.41 | 79.01 | −0.168 |
| CsaV3_3G001740.1 | PYL3 | 3 | 1311481 | 1312909 | 229 | 25.70 | 5.41 | 49.19 | 80.87 | −0.519 |
| CsaV3_3G008140.1 | PYL4 | 3 | 7005104 | 7008146 | 185 | 20.81 | 8.28 | 39.57 | 87.84 | −0.391 |
| CsaV3_3G022000.1 | PYL5 | 3 | 19093676 | 19095745 | 184 | 20.58 | 5.97 | 47.35 | 100 | −0.217 |
| CsaV3_3G033450.1 | PYL6 | 3 | 28742374 | 28744464 | 206 | 22.59 | 6.44 | 48.23 | 85.05 | −0.274 |
| CsaV3_3G049190.1 | PYL7 | 3 | 40090988 | 40092469 | 232 | 25.47 | 5.35 | 32.61 | 78.97 | −0.306 |
| CsaV3_4G026530.1 | PYL8 | 4 | 15776156 | 15778129 | 233 | 25.66 | 7.73 | 46.6 | 81.93 | −0.359 |
| CsaV3_4G035560.1 | PYL9 | 4 | 25042643 | 25043922 | 193 | 21.59 | 5.47 | 41.14 | 81.76 | −0.473 |
| CsaV3_5G000620.1 | PYL10 | 5 | 319763 | 324420 | 195 | 21.89 | 6.44 | 36.06 | 94.87 | −0.423 |
| CsaV3_5G008890.1 | PYL11 | 5 | 5494966 | 5495652 | 179 | 19.90 | 4.91 | 34.88 | 84.92 | −0.158 |
| CsaV3_5G010560.1 | PYL12 | 5 | 6531332 | 6532063 | 243 | 25.86 | 6.79 | 50.96 | 82.22 | −0.187 |
| CsaV3_6G001990.1 | PYL13 | 6 | 1361372 | 1362257 | 208 | 23.02 | 6.59 | 41.55 | 90.19 | −0.067 |
| CsaV3_7G025250.1 | PYL14 | 7 | 14609356 | 14611908 | 236 | 26.69 | 6.66 | 43.33 | 94.07 | −0.088 |
| Gene ID | Gene name | α-helix % | Beta turn % | Random coil % | Extended strand % | Subcellular localization |
|---|---|---|---|---|---|---|
| CsaV3_2G030780.1 | PYL1 | 37.04 | 6.17 | 38.27 | 18.52 | Cytoplasm, Nucleus |
| CsaV3_2G030790.1 | PYL2 | 30.23 | 8.72 | 39.53 | 21.51 | Cytoplasm, Nucleus |
| CsaV3_3G001740.1 | PYL3 | 38.43 | 5.68 | 39.74 | 16.16 | Cytoplasm, Nucleus |
| CsaV3_3G008140.1 | PYL4 | 39.46 | 5.41 | 37.30 | 17.84 | Cytoplasm |
| CsaV3_3G022000.1 | PYL5 | 41.30 | 5.98 | 35.87 | 16.85 | Cytoplasm |
| CsaV3_3G033450.1 | PYL6 | 29.13 | 6.80 | 43.20 | 20.87 | Cytoplasm, Chloroplast |
| CsaV3_3G049190.1 | PYL7 | 40.95 | 8.19 | 35.34 | 15.52 | Cytoplasm |
| CsaV3_4G026530.1 | PYL8 | 35.9 | 3.85 | 44.02 | 16.24 | Cytoplasm, Chloroplast |
| CsaV3_4G035560.1 | PYL9 | 40.41 | 5.18 | 35.23 | 19.17 | Nucleus, Cytoplasm |
| CsaV3_5G000620.1 | PYL10 | 36.41 | 4.62 | 41.54 | 17.44 | Nucleus, Cytoplasm |
| CsaV3_5G008890.1 | PYL11 | 46.37 | 3.91 | 31.84 | 17.88 | Nucleus |
| CsaV3_5G010560.1 | PYL12 | 35.8 | 7.00 | 40.33 | 16.87 | Chloroplast |
| CsaV3_6G001990.1 | PYL13 | 29.81 | 6.25 | 39.90 | 24.04 | Cytoplasm, Chloroplast |
| CsaV3_7G025250.1 | PYL14 | 42.37 | 3.39 | 34.75 | 19.49 | Cytoplasm, Chloroplast |
Subcellular localization prediction display (Table 2) that most of the PYL genes in cucumber were predicted to be located in cytoplasm and nucleus, while CsPYL6, CsPYL8, CsPYL12, CsPYL13 and CsPYL14 were located in chloroplast. The results of secondary structure prediction (Table 2) found that the secondary structure of most CsPYL proteins were mainly composed of α-helix and irregular curl, and the proportion of extended structure and β-angle were the smallest.
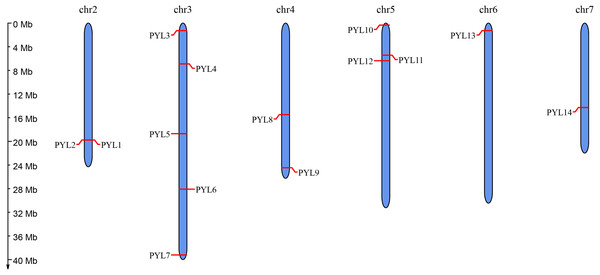
Moreover, to predict the position of CsPYLs gene on the chromosome information, using the online website MG2C to map the chromosomal location. The distribution of PYL genes on chromosomes showed that 14 members of PYL gene family were distributed on 6 chromosomes of cucumber (Fig. 1). There were more PYL genes on chromosomes 3 (PYL3-7) and 5 (PYL10-12). Both CsPYL1 and CsPYL2 were located on the same site of Chr 2 and belonged a pair of tandem duplication genes.
Collinearity analysis of the PYL gene family in cucumber
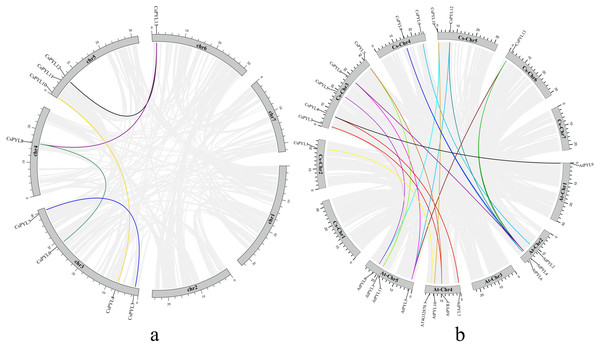
Using Mcscanx and TBtools to analyze the segmental duplication events, it was found that there were 5 pairs of collinearity genes in cucumber (Fig. 2A), including 4 segmental duplication events between different chromosome, containing (CsPYL4/CsPYL10, CsPYL6/CsPYL8, CsPYL8/CsPYL13, CsPYL12/CsPYL13), the other 1 duplication events within the same chromosome, including (CsPYL3/CsPYL7).
Subsequently, we also explored the collinearity relationships between the cucumber PYL genes and Arabidopsis (Fig. 2B). There were 19 collinear gene pairs between 11 CsPYLs and 11 AtPYLs. According to the collinearity relationship between CsPYLs and AtPYLs, most collinear gene pairs were distributed on At-Chr 2, 4, 5 and Cs-Chr 3-5. In addition, the collinear relationship between most CsPYLs and AtPYLs was one-to-many, such as the collinear relationships between AtPYL1 and CsPYL3, 7; AtPYL4 and CsPYL6, 8, 13. The genes with collinear relationship in our study belong to the same group in the phylogenetic tree (Fig. 3) and were very similar to their gene structure and conserved motifs (Fig. 4), such as AtPYL1, AtPYR1 and CsPYL3, 7 which all have upstream/downstream sequence and two exons (except CsPYL3).
Figure 1: Chromosomal distribution and localization of CsPYLs.
The chromosome names are shown at the top of each chromosome. The chromosome scale is in millions of bases (Mb) on the left.Figure 2: Collinearity analysis of the PYL genes family.
(A) Collinearity analysis of the PYL genes family in cucumber. Chromosomes 1–7 are represented by gray rectangles. The gray lines indicate synteny blocks in the cucumber genome, while the lines of different colors between chromosomes delineate segmental duplicated gene pairs. (B) Collinearity analysis of the PYL gene family between cucumber and Arabidopsis thaliana. Gray lines denote the collinear blocks between cucumber and Arabidopsis genomes and thelines of different colors denote the syntenic gene pairs of PYLs. Gray rectangles represent respectively the cucumber chromosomes (1–7) and Arabidopsis chromosomes (1–5).Analysis of dN/ds values of PYLs in cucumber, cucumber and Arabidopsis
To further investigate the divergence and selection in duplication of PYL genes, the non-synonymous substitution rate (dN), synonymous substitution rate (ds) and dN/ds values were evaluated for the homologous gene pairs among cucumber, cucumber and Arabidopsis (Table 3). When dN/dS > 1 is the positive selection, dN/d S = 1 is the neutral selection, 0 < dN/dS <1 is purifying selection (Yadav et al., 2015). The dN/ds value of all cucumber gene pairs was less than 1. Similarly, the dN/ds value of all collinear gene pairs in cucumber and Arabidopsis was less than 1. These data suggest that these genes were mainly under the purifying selection during evolution and could help to maintain the basic function of this gene.
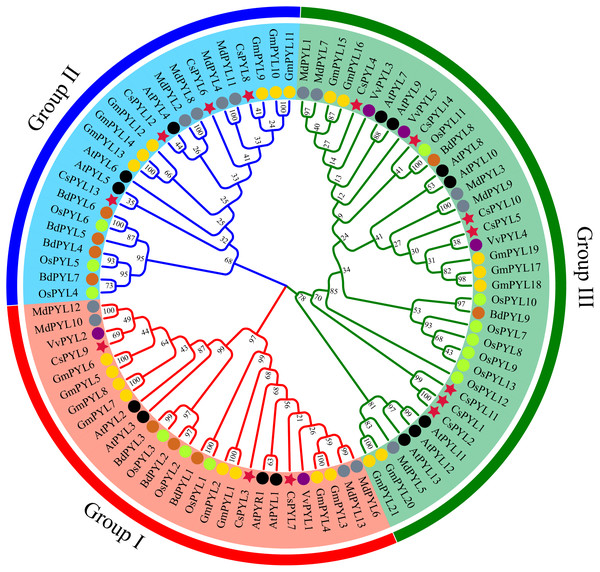
Figure 3: PYL phylogenetic trees of seven plants.
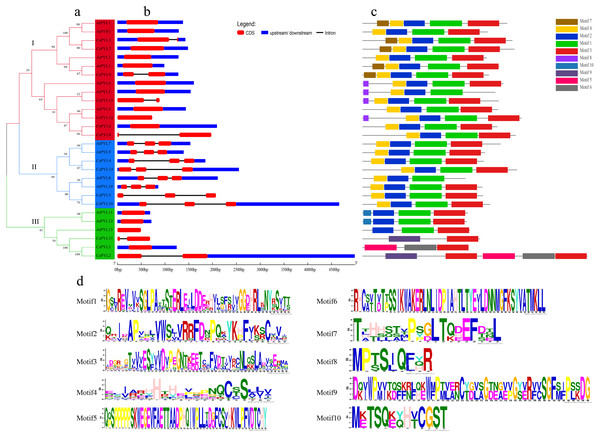
Red circles represent C. sativus, black circles represent A. thaliana, blue circles represent M. domestica, green circles represent O. sativa, yellow circles represent G. max, purple circles represent V. vinifera, and pink circles represent B. distachyon.Figure 4: Phylogenetic relationships, gene structure and conserved motifs of PYL genes.
(A) Phylogenetic relationship of PYLs from in Arabidopsis and cucumber. Tree was constructed by the Maximum likelihood method. (B) Exon/intron architectures of PYLs. Red colour boxes indicate exons and lines represent introns. (C) Distributions of conserved motifs in PYL proteins. Motifs are indicated by 10 different colour boxes. (D) The pattern identification of ten conservative sequences.Phylogenetic analysis of the CsPYLs family
In order to determine the evolutionary relationship of PYL genes, a phylogenetic tree was constructed using the 14 CsPYL genes from C. sativus, 14 AtPYLs from A. thaliana, 13 OsPYLs from O. sativa, 21 GmPYLs from G. max, 5 VvPYLs from V. vinifera, 9 BdPYLs from B. distachyon and 13 MdPYLs from M. domestica (Supplemental Information 4). A total of 89 PYL genes of the five plants were divided into three groups (Fig. 3), namely group I-III, which consisted 27, 24 and 38 members respectively. Every group contained PYLs from eudicots and monocots, hinting that these PYLs generated before the divergence from eudicots and monocots.
| species | A pair of genes | S | N | ds | dN | dN/ds |
|---|---|---|---|---|---|---|
| cucumber | CsPYL3-CsPYL7 | 177.3 | 482.7 | 3.558 | 0.2446 | 0.0688 |
| CsPYL6-CsPYL8 | 136 | 470 | 57.1029 | 0.2383 | 0.0042 | |
| CsPYL4-CsPYL10 | 137.7 | 417.3 | 9.8761 | 0.1667 | 0.0169 | |
| CsPYL12-CsPYL13 | 156.5 | 389.5 | 44.7183 | 0.2544 | 0.0057 | |
| CsPYL8-CsPYL13 | 141.3 | 413.7 | 50.1652 | 0.3002 | 0.006 | |
| cucumber andArabidopsis | AtPYL1-CsPYL3 | 170.2 | 474.8 | 48.4185 | 0.3046 | 0.0063 |
| AtPYL1-CsPYL7 | 168.7 | 485.3 | 49.5664 | 0.2856 | 0.0058 | |
| AtPYL2-CsPYL9 | 127.7 | 436.3 | 2.3335 | 0.2376 | 0.1018 | |
| AtPYL4-CsPYL6 | 138.6 | 470.4 | 56.0232 | 0.3216 | 0.0057 | |
| AtPYL4-CsPYL8 | 136 | 473 | 6.4985 | 0.2768 | 0.0426 | |
| AtPYL5-CsPYL6 | 139.1 | 430.9 | 52.2168 | 0.3448 | 0.0066 | |
| AtPYL5-CsPYL12 | 163.2 | 442.8 | 47.2935 | 0.3561 | 0.0075 | |
| AtPYL5-CsPYL13 | 133.2 | 400.8 | 51.1961 | 0.3025 | 0.0059 | |
| AtPYL6-CsPYL8 | 127.2 | 472.8 | 60.1089 | 0.3307 | 0.0055 | |
| AtPYL6-CsPYL12 | 167.2 | 474.8 | 48.8969 | 0.3593 | 0.0073 | |
| AtPYL6-CsPYL13 | 140.1 | 408.9 | 48.2391 | 0.2968 | 0.0062 | |
| AtPYL7-CsPYL4 | 130.4 | 424.6 | 3.572 | 0.1962 | 0.0549 | |
| AtPYL8-CsPYL5 | 139.2 | 412.8 | 2.3447 | 0.1336 | 0.057 | |
| AtPYL8-CsPYL10 | 122.4 | 441.6 | 3.7946 | 0.1269 | 0.0334 | |
| AtPYL9-CsPYL4 | 126.9 | 416.1 | 3.1501 | 0.1713 | 0.0544 | |
| AtPYL10-CsPYL10 | 129.2 | 416.8 | 2.4261 | 0.1965 | 0.081 | |
| AtPYR1-CsPYL3 | 150.1 | 416.9 | 25.5091 | 0.2497 | 0.0098 | |
| AtPYR1-CsPYL7 | 152.3 | 420.7 | 48.2234 | 0.2499 | 0.0052 |
Gene structure and conserved domains analysis of CsPYLs
To understand the structure of the cucumber PYL gene family, we analyzed the phylogenetic relationship of the CsPYL and AtPYL proteins (Fig. 4A), gene structure (Fig. 4B) and conserved motif (Fig. 4C). The 14 CsPYL genes were divided into three groups. In the group I, all the PYL genes included one or two exons. CsPYL3, 9, 13 and 8 have two exons, and both have one intron with different lengths, among which CsPYL8 and 13 have no upstream and downstream gene sequences. In the group II, all PYL genes have three exons and two introns, only CsPYL5 has no upstream and downstream sequences. In the group III all PYL genes have one or two exons, only AtPYL13 and CsPYL11 has no non-coding regions. CsPYL12 had the longest fragment length that was more than 4,500 bp.
The protein sequences of CsPYLs and AtPYLs were analyzed by MEME website, and a total of 10 conserved motifs were identified (Fig. 4C). All the PYL members contain 2 to 5 conserved motifs. Compared with other genes, CsPYL genes in group III had no motif 1 and 2, while AtPYL8 and AtPYL5 did not have motif 3. Except all PYL genes in the group III other PYL genes have motif 4. This indicates that PYL proteins have highly conserved amino acid residues. Besides the common motif, PYL genes in each group have its own unique motif, such as AtPYL1, CsPYL7, CsPYL3, AtPYL2, CsPYL9, AtPYL3 in group I contain motif 7. CsPYL1, CsPYL2 in group III contained motif 5, motif 6 and motif 9 which were not found in other groups. From these, the proteins classified into the same group share similar motif characteristics, suggesting functional similarities among members, while presence of unique motifs might carry out unique/specialized biological functions.
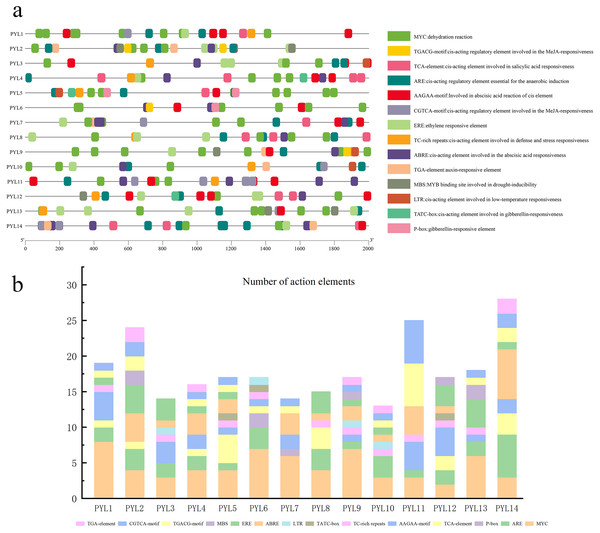
Cis-Element analysis of the CsPYLs promoter in cucumber
The 2,000 bp sequence before the start codon was selected to predict cis-acting elements. As is shown in Fig. 5A, and it was found that the regulatory elements of the CsPYLs were very abundant in number and variety, mainly analyzes the hormone regulation and stress-related elements. Among the 14 CsPYLs promoters, contain 4 stress-related elements: MYC, TC-rich repeats, MBS, LTR. Additionally, there were 6 hormone-related cis-acting elements predicted, for instance, CGTCA-motif and TGACG-motif are related to MeJA responsive element; TATC-box and P-box are related to Gibberellin-responsiveness responsive element; AAGAA-motif and ABRE are related to abscisic acid response element; TCA-element are related to SA responsive element, as well as ethylene response element (ERE) and auxin response element (TGA-element).
Figure 5: Putative cis-elements existed in the 2 kb upstream region of cucumber PYL genes.
(A) The elements which respond to hormones are displayed in differently coloured boxes. The homeopathic elements represented by different color boxes and their names and functions. (B) Cucumber PYL gene cis-acting elements and number statistics.The promoter regions of 14 CsPYL genes contain at least 6 acting elements, and all CsPYL gene contains MYC and ARE elements (except for CsPYL7), among which MYC has the largest number. It can be seen from the statistical table (Fig. 5B) that CsPYL14 has the largest number of cis-acting elements, followed by CsPYL2 and CsPYL11.
Tissue-specific expression of CsPYL genes in cucumber
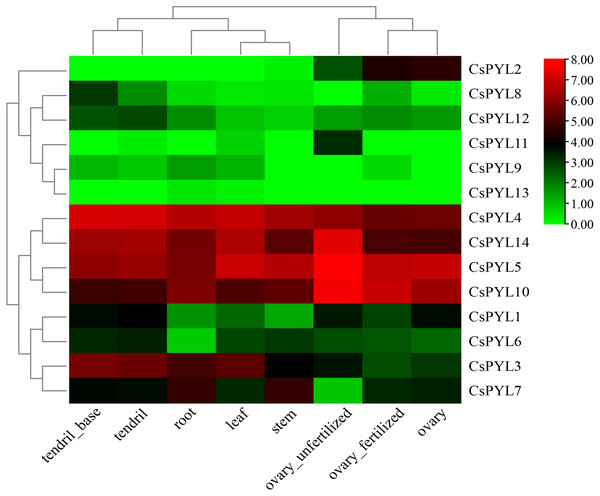
In order to better understand the role of CsPYL genes in cucumber growth and development, transcriptome data from cucumber database were used to analyze their expression in different tissues. Expression patterns of 14 CsPYL genes in different tissues were constructed based on transcriptome data (Fig. 6). The expression of CsPYL1, CsPYL3-7, CsPYL10 and CsPYL14 in different eight tissues were high. On the contrary, the expression levels of CsPYL9, CsPYL11 and CsPYL13 in tissues were very low, and CsPYL13 was almost not detected, while CsPYL11 is only elevated in unfertilized ovaries. The expression of CsPYL2 was high in the fertilized ovaries, unfertilized ovaries and ovary, but even hardly detected in other parts. The spatial variations in the expression of CsPYL genes in different tissues indicate that they may participate in different stages of cucumber growth and development processes.
Figure 6: Tissue-specific digital expression profifiles of the CsPYL genes in cucumber.
The CsPYL genes were listed at the right of the expression array, and the expression values mapped to a color gradient from low (green) to high expression (red) are shown at the right of the figure.Response of CsPYL genes expression to various abiotic stresses and ABA treatment
PYLs, as ABA receptors, might play a vital function in abiotic stress signaling pathways. To further understand the expression of PYL genes under different abiotic stresses, qRT-PCR was used to analyze the expression of 14 cucumber CsPYL genes upon various abiotic stress treatments (including ABA, NaCl, PEG). Raw CT values were placed in (Supplemental Information 5).
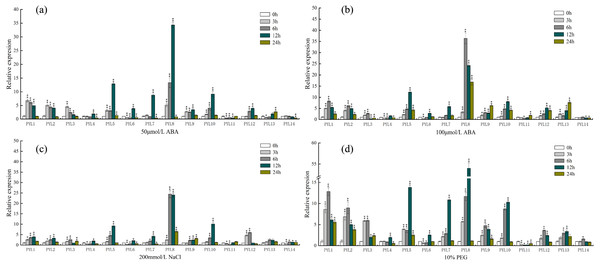
As shown in Fig. 7A, under the treatment of 50 µmol/L ABA, the expression of PYL8 was up-regulated at all time points (except 24 h), and reached the highest level at 12 h, which was 30 times higher than that of 0 h (control check). Compared to the 0 h, the relative expression quantity of PYL1-3, PYL5, PYL9 and PYL10 were up-regulated about 3 h, 6 h and 12 h. However, after 24 h, the expression level of this genes was close to 0 h. On the Contrary, compared with 0 h, PYL11 relative expression was down-regulated about 3 h, 6 h and 12 h, then increased slowly, and reached the 0 h level at 24 h. Expression of PYL14 continually decreased in response to 50 µmol/L ABA. When ABA concentration doubled (Fig. 7B), the expression levels of PYL1, PYL2, PYL5, PYL8-10, PYL12 and PYL13 were all up-regulated in the four time periods, especially PYL8 at 24 h, which was 36 times higher than 0 h. PYL11 showed the same changes as 50 µmol/L ABA treatment, but decreased slightly at 3 h, 6 h and 12 h, and up-regulated at 24 h, which was 1.8 times of 0 h. Only the expression of PYL14 was slightly down-regulated at all time points.
Figure 7: Expression profiles of CsPYL genes in response to various stress.
(A) A total of 50 µmol/L ABA, (B) 100 µmol/L ABA, (C) 200 mmol/L NaCl, and (D) 10% PEG . The relative expression levels of CsPYL genes were analyzed at 3, 6, 12 and 24 h of treatments as compared with their values at 0 h. The gene relative expression was calculated using the 2−ΔΔCt method with CsActin as an internal control, and value represents mean ±SE of three biological replicates. Asterisks indicated values that are significantly different from CK (0 h) (* p < 0.05, ** p < 0.01, one-way ANOVA).After 200 mmol/L NaCl treatment for (Fig. 7C) 12 h, the relative expression levels of PYL5, PYL8 and PYL10 up-regulate and were 9, 24 and 10 times higher than that of 0 h respectively. Compared with 0 h, except PYL4, PYL6 and PYL11 were down-regulated at 3 h and 6 h, other genes were up-regulated in varying degrees. The relative expression of PYL14 was basically unchanged. Most of the genes were up-regulated after 10% PEG (Fig. 7D), especially PYL8 at 12 h, which was 53 times higher than 0 h. It can be seen that the relative expression of PYL genes under 10% PEG treatment is higher than other treatments, such as PYL1-5. This result showed that many CsPYL genes (especially PYL8) were likely to play critical roles in the abiotic and hormonal stress signaling transduction pathways.
Discussion
Studies show that ABA can not only regulate the growth and development of plants, but also improve the stress resistance of plants. ABA is perceived by PYL family of receptors, which are the largest plant hormone receptor family known (Dupeux et al., 2011), PYLs play an important role in ABA signal transduction. In our study, we found 14 CsPYL genes, which was almost equal number of AtPYL (Park et al., 2009) from Arabidopsis. Chromosome mapping showed that (Fig. 1) PYL genes distributed on 6 chromosomes, and most of them were distributed on chromosome 3, and there was no clustering phenomenon. However, PYL gene of rice and Brassica napus were partially clustered (Di et al., 2018; Yadav et al., 2020), which may indicate that there were different trajectories in the evolution of different plants. Segmental duplication, tandem duplication and transposition events are the main reasons for gene family expansion (Kong et al., 2007). Synteny analysis suggested that most CsPYL genes were involved in syntenic blocks (Fig. 2A), indicating that segmental duplication events play major roles in the expansion of the CsPYLs in cucumber. The collinear relationship (Fig. 2B) between cucumber and A. thaliana PYL genes was analyzed, and 20 pairs of collinear genes were found between cucumber and A. thaliana. Also, the dN/dS values of these gene replication pairs were all less than 1, indicating that they had been purified and selected to predict their high conservatism in the evolution process (Table 3).
According to the phylogenetic tree analysis (Fig. 3), PYL genes of C. sativus, A. thaliana, O. sativa, G. max, V. vinifera, B. distachyon and M. domestica were divided into three groups, which were consistent with the grouping of rice (Yadav et al., 2020), Brassica napus (Di et al., 2018), cotton (Zhang et al., 2017) and rubber trees (Guo et al., 2017), but were different from the groupings of the apple which were divided into four groups (Hou et al., 2020). This difference may be due to the diversity of different species sequences. Moreover, we observed cucumber PYLs clustered more closely with dicotyledonous plants PYLs than monocotyledonous plants, in agreement with the evolutionary relationships among these plants.
Meanwhile, Fig. 4A showed that the groups of the cluster analysis of CsPYL and AtPYL also had three. The exon/intron structure of genes is an important marker to reveal the evolutionary relationship between members of the gene family (Long et al., 2003). The gene structure (Fig. 4B) showed that that all CsPYL genes have one to three exons, only CsPYL1, CsPYL6 and CsPYL7 have one exon, and they all have two non-coding regions and no introns. The members in different groups are similar in structure, some closely related PYL genes have similar lengths of exons, such as all members in group II, suggesting that these genes are highly conserved during evolution. The conserved domains of CsPYL and AtPYL genes analysis showed that motif 3 was found in all PYL genes (except AtPYL8, 5). Besides CsPYL2, CsPYL1 and CsPYL11 in group III, motif 1 and motif 2 were found in all PYL genes. This indicates that motif 1, 2 and 3 are highly conserved. HHpred analysis to affirm if the motifs obtained from the MEME analysis are similar to any of the known protein motifs (Table S2), it was found that all three motifs belong to the Polyketide cy-clase/dehydrase family, which are enzymes involved in polyketide synthesis (Iyer, Koonin & Aravind, 2001). It was observed that these novel motifs did not show any significant similarity with the known motifs, which was consistent with the findings in rice (Yadav et al., 2020). This also indicates that all the identified CsPYLs have typical subfamily features and the proteins classified into the same subgroup share similar protein motifs.
Understanding the protein’s subcellular location information (Table 2) may provide us with the necessary help to infer the biological function of the protein, CsPYLs would mainly locate in nucleus, cytoplasm and chloroplast, so it was speculated to be related to photosynthesis, respiratory action and cell growth and development. The plant tissue expression specificity (Fig. 6) showed that PYL genes had varying degrees tissue-specific expression. In rubber trees, Brassica napus and rice PYL genes also show preferential expression in different tissue types. (Guo et al., 2017)(Di et al., 2018)(Yadav et al., 2020). Our study showed that only CsPYL13 and CsPYL9 were low or not expressed in all tissues. The expression levels of CsPYL2 were higher in fertilized ovary, unfertilized ovary and ovary, CsPYL8 and CsPYL12 were higher in the tendril base, and CsPYL11 were higher in unfertilized ovary, but the expression levels of these genes were lower in other organs. Other genes, such as CsPYL4, 5, 10, and 14 were highly expressed in all tissues. In a previous study by Wang et al. (2012b), their research found that, the CsPYL2 may involve in transducing ABA signal in fruit and regulating fruit development and ripening. Under drought stress condition in cucumber seedlings. CsPYL1 and CsPYL2, were sensitive and up-regulated in root, stem and leaf; CsPYL3 showed a low sensitivity and were down-regulated in root and stem. In our study, CsPYL10, CsPYL13 and CsPYL14 correspond to PYL1, PYL3 and PYL2 in the study of Wang et al., and CsPYL10, CsPYL14 were highly expressed in different tissues of cucumber. On the contrary, CsPYL13 was almost not expressed in different tissues, which was consistent with their study.
When plants are under various stresses, these stresses signals will activate transcription factors, which combine cis-acting elements to stimulate the expression of related genes. (Mishra et al., 2014; Xing et al., 2018). In plant development and resistance to various stresses, cis-acting elements are important regulators of hormone response (Nishimura et al., 2010). In this study, among all the functional elements, MYC element exists in every CsPYL gene, and drought responsive elements are also abundant in PYL genes of rubber trees and cotton (Zhang et al., 2017; Guo et al., 2017). The results of qRT-PCR (Fig. 7D) suggested that CsPYLs expression were higher than 0 h with 10% PEG treatment (Except PYL4, 11, 14), indicating that most of CsPYLs may had drought resistance, which was similar to the results of cis-acting element. In addition, the CsPYLs also contained other hormone related cis-acting elements, such as a large number of ABA response sites (AAGAA-motif, ABRE).
In Arabidopsis, over-expression of PYL4 and PYL9 can enhance the drought resistance (Pizzio et al., 2013; Zhao et al., 2016)[18,19]. From the phylogenetic tree (Fig. 3), AtPYL4 and CsPYL12 are in the same group, AtPYL9 and CsPYL14, CsPYL4 in the same group. After different treatments (Fig. 7), CsPYL14 and CsPYL4 were almost not expressed, while CsPYL12 was up-regulated, but it was not obvious compared with other genes. This suggests that the relative expression levels of PYL gene may vary among different species. Our study shows that, under 50 µmol/LABA treatment (Fig. 7A), the expression levels of CsPYL5, 7, 8 and 10 were up-regulated for 3 h, 6 h and 12 h relative to 0 h, and CsPYL8 up-regulation is most obvious. When the concentration of ABA increased (Fig. 7B), the relative expression of some genes were up-regulated at 24 h, such as PYL1, PYL2, PYL5, PYL8-10, especially PYL8, which was more 10 times than 0 h. It was speculated that when ABA concentration increased, the expression level of PYL genes was affected by ABA more lasting. Overall, except PYL4, PYL11 and PYL14, all CsPYL genes increased more or less under ABA treatment. This was similar to the result of apple PYL genes under ABA treatment (Hou et al., 2020), the expression of all MdPYLs except for MdPYL11 exhibited significant increases by ABA treatment. However, some GhPYL genes were down-regulated under ABA treatment in cotton. After treatment with ABA, GhPYR1-1A, GhPYR1-3D, GhPYL9-5A significantly reduced, and GhPYL9-2D was not influenced by ABA treatment (Zhang et al., 2017; Chengzhen et al., 2017). This also indicates that PYL genes in different crops have different responses to ABA during the evolutionary process and a great number of CsPYLs have different responses to ABA. Under salt stress (Fig. 7C) and drought stress (Fig .7 d) CsPYL5, 8, 10 were obviously up-regulated and the relative expression of PYL genes with 10% PEG treatment is higher than other treatments, such as PYL1-5 and PYL8. But in rice, except for OsPYL4, all PYLs were down-regulated under drought stress and OsPYL12 was unaltered. In Arabidopsis, overexpression of AtPYL4, 5, 7, 8 and 13 respectively enhanced drought tolerance (Lee et al., 2015; Yadav et al., 2020; Zhao et al., 2016). However, in the study of duodecuple mutants PYR1 and PYL1-12 by Zhao et al. (2018), it was found that the duodecuple mutant was extremely insensitive to ABA effects on seedling growth, stomatal closure, leaf senescence, and gene expression. Therefore, PYL genes may have a potential effect on improving abiotic stress tolerance in plants. Together, all these results show that many CsPYLs were likely respond to ABA, drought, salt treatments, and CsPYL genes respond more significantly to drought stress than other treatments.
Conclusion
We identified 14 PYL genes in cucumber, which were distributed on 6 chromosomes, and more members on chromosome 3 (PYL3-7) and 5 (PYL10-12). Motif 3 was found in all CsPYL genes, and its structure was conserved. The analysis of cis-acting elements showed that there were many elements in cucumber PYL gene responding to abiotic stress and hormones, and drought responsive element MYC was present in all CsPYL genes. Furthermore, qRT-PCR analysis showed that CsPYLs (especially PYL8) may have certain function in resisting abiotic and hormone stresses. This study provided relevant information for the study of PYL gene function in cucumber.