The role of CD47 in immune escape of colon cancer and its correlation with heterogeneity of tumor immune microenvironment
- Published
- Accepted
- Received
- Academic Editor
- Brittany Lasseigne
- Subject Areas
- Bioinformatics, Cell Biology, Gastroenterology and Hepatology, Oncology
- Keywords
- CD47, Immune escape, Tumor microenvironment, COAD, Heterogeneity
- Copyright
- © 2024 Tian et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits using, remixing, and building upon the work non-commercially, as long as it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2024. The role of CD47 in immune escape of colon cancer and its correlation with heterogeneity of tumor immune microenvironment. PeerJ 12:e18579 https://doi.org/10.7717/peerj.18579
Abstract
Background
Cluster of differentiation 47 (CD47), a transmembrane protein, plays a critical role in regulating cellular functions and maintaining immune homeostasis. Its expression has been shown to influence cancer prognosis. In this study, we investigated the role of CD47 in tumor progression in colon adenocarcinoma (COAD) and evaluated its potential as a target for immunotherapy.
Materials and Methods
We analyzed surgical samples from 96 COAD patients. Immunohistochemical analysis was performed on 90 samples, while the remaining 6 were subjected to multiplex immunofluorescence. To explore the association between CD47 expression and clinicopathological characteristics, we integrated transcriptome data from The Cancer Genome Atlas and the Gene Expression Omnibus using R software. The Tumor Immune Estimation Resource and Kaplan-Meier plotter were utilized to assess the relationship between CD47 expression, patient prognosis, and immune infiltration. Furthermore, the single-cell Tumor Immune System Interaction Database was used to examine the correlation between CD47 expression and the tumor microenvironment (TME). All included patients gave oral and written informed consent. The study was approved by the Ethics Committee of 3201 Hospital (full name: Medical Ethics Committee of 3201 Hospital).
Results
CD47 was found to be overexpressed in various tumors, including COAD. Higher CD47 expression was significantly associated with more advanced tumor stages, including TNM staging, T staging, and N staging (P < 0.05). A robust correlation was observed between CD47 expression and immune cell infiltration in COAD. Patients with elevated CD47 expression tended to have longer disease-free intervals, although this benefit was diminished in cases with high infiltration of M1 macrophages. The immunosuppressive function of CD47 primarily acted through the CD47/SIRPα pathway. Additionally, distinct cellular compositions and distributions were identified between primary and metastatic COAD, underscoring the heterogeneity of the TME. CD47 also influenced the TME by modulating cytokine and cytokine receptor interactions.
Conclusion
CD47 represents a promising prognostic biomarker and a potential target for immunotherapy in COAD. These findings provide new insights into therapeutic strategies aimed at modulating the TME and improving patient outcomes.
Introduction
Colorectal cancer (CRC) is the fourth most common cancer globally and the third leading cause of cancer-related deaths. Among its subtypes, colon adenocarcinoma (COAD) is the most prevalent, accounting for over 60% of all CRC cases (Bray et al., 2018). COAD is characterized by a complex interplay of genetic, cellular, and environmental factors, which contribute to its high incidence and mortality rates (Brenner, Kloor & Pox, 2014). Understanding the underlying mechanisms driving COAD development and identifying reliable biomarkers are essential for improving patient outcomes.
CD47, a cell surface protein belonging to the immunoglobulin superfamily, is widely expressed across various cell types and is notably overexpressed in many tumors. By interacting with signal regulatory protein alpha (SIRPα) on macrophages, CD47 transmits a “don’t eat me” signal, enabling tumor cells to evade immune surveillance (Logtenberg, Scheeren & Schumacher, 2020; Zhao et al., 2016; Cioffi et al., 2015; Klimp et al., 2002). As an innate immune checkpoint, CD47 overexpression has been associated with poor survival outcomes in several types of cancer. In CRC, intrinsic CD47 signaling has been linked to increased glycolysis, thereby enhancing tumor growth and metastasis (Hu et al., 2020). Moreover, CD47 plays a role in regulating tumor cell apoptosis and angiogenesis in CRC (Oh et al., 2022). In Epstein-Barr virus-associated gastric cancer, CD47 has been shown to suppress anti-tumor immunity by limiting T-cell infiltration (Duan et al., 2023). Despite these findings, the specific role of CD47 in COAD, particularly its influence on tumor immunity, remains poorly understood and warrants further investigation. Studies have shown that decreasing CD47 expression increases CD8+ T cell infiltration and enhances anti-tumor immune responses (Zhao et al., 2024). Multiple factors in the tumor microenvironment (TME), including immunosuppressive cells and metabolic reprogramming, affect CD47 function. Cancer cells adapt to the TME by altering their metabolic state, which further enhances the immunosuppressive effects of CD47 (Van Weverwijk & De Visser, 2023). Due to the key role of CD47 in tumor immune escape, blocking the interaction of CD47 with SIRPα has become a new therapeutic strategy. Immune checkpoint inhibitors (ICBs) and other immunotherapies are currently under development to enhance the immune system’s ability to recognize and attack tumors (Hu et al., 2024a; Hu et al., 2024b). The exact impact of CD47 in COAD, particularly concerning tumor immunity, requires further investigation.
Materials and Methods
Clinical data collection and screening
A total of 96 COAD samples were collected between January 2021 and July 2022 at Jiangjin Central Hospital Chongqing University. The study was approved by the hospital’s Ethics Committee (approval number: KY2022037, TYYLKYJJ-2023-014), and all participants provided written informed consent prior to inclusion.
The inclusion criteria for the study were: patients with a first-time diagnosis of COAD and no prior history of related malignancies or previous surgeries. (1) Patients with no previous gastrointestinal endoscopy and initial colonoscopy biopsy diagnosed with COAD; (2) Patients with previous gastrointestinal endoscopy, history of colon polyps and regular follow-up progression to colon cancer. All patients underwent histopathological confirmation of COAD. Detaild clinical characteristics of the participants are presented in Table S1.
Immunohistochemistry (IHC)
All COAD samples underwent standard histopathological processing, including fixation in 10% neutral formalin, dehydration, paraffin, embedding, and sectioning at a thickness of 3 µm. The sections were dried and pretreated using the DAKO PT Link automated immunohistochemical preconditioning system. Immunohistochemical staining was carried out with the DAKO AutostainerLink 48 automated system, employing the EnVision method. The primary antibody used was anti-CD47 (1:200, CST, 63000S). Tonsillar tissue was utilized as a positive control for external validation, while phosphate-buffered saline (PBS) was used as a negative control in place of the primary antibody. In addition, red blood cells within the blood vessels served as an internal control.
Interpretation of the IHC results
CD47 expression was predominantly localized to the cell membrane and cytoplasm of tumor cells. To assess CD47 staining in COAD samples, two independent and experienced pathologists performed a double-blind evaluation under high-power microscopy (400x magnification). The staining was scored based on both the intensity and the proportion of positive cells. Staining intensity for the cell membrane and cytoplasm was graded on a scale of 0 to 3: 0 points for no staining, 1 point for 1–25% positive staining, 2 points for 26–50%, and 3 points for 51–100%.
CD47 expression and the clinical dataset
Data on CD47 mRNA expression and clinical information for COAD patients were retrieved from The Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/). These data were processed using R software (v3.6.3) and Perl(version 5.3.0), and the deadline was March, 2023. The dataset included 480 COAD patients, of whom 41 had both normal and cancerous colon tissue samples, while the remaining 439 samples contained only cancerous tissue. Patients were divided into high and low CD47 expression groups based on the median CD47 mRNA expression level. ROC analysis of the data was performed using ROC (1.18.0) and the results were visualized with ggplot2 (3.3.6), correcting the outcome order of the data by default (ensuring the results are upward convex).
In addition, The GSE33113 dataset was obtained from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/) database via GEOquery (2.64.2). GSE33113 was located on the GPL570 platform ((HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array). This dataset comprised primary tumors from 90 stage II CRC patients, along with 6 matched normal colon tissue samples. From this dataset, From this dataset, CD47 gene expression data were extracted for analysis. The study design and human data used in this analysis are presented in Table S2.
Survival analysis and clinical correlation analysis
The samples were stratified into high and low expression groups based on median CD47 gene expression levels. Kaplan–Meier survival curve analysis, along with data from the GEPIA database, was used to evaluate the correlation between CD47 expression and survival outcomes in COAD patients. Additionally, the association between CD47 expression, clinical parameters, and patient prognosis was further investigated using univariate and multivariate Cox regression analyses. Hazard ratios (HR) were calculated to assess the relative risk linked to CD47 expression. The multivariate or univariate Cox regression models were performed using R package “survival”. Logistic regression analysis was used to explore the correlation of CD47 expression with clinical factors of COAD using the R package “Limma”.
Multiplex immunofluorescence staining
Multiplex immunofluorescence staining was performed according to previously published protocols. The primary antibodies used included anti-CD47 (rabbit, 1:200, CST, 63000S), anti-CD68 (rabbit, 1:3000; AiFang Biological, Changsha, China), and anti-CD163 (rabbit, 1:3000, Proteintech, China). Signals were detected using horseradish peroxidase-conjugated secondary antibodies (GB23301, GB23303; Servicebio, Beijing, China), followed by tyramide signal amplification with FITC-TSA, CY3-TSA, 594-TSA, and 647-TSA (Servicebio). Multispectral images were acquired and analyzed using CaseViewer (C.V 2.3, C.V 2.0) and Panoramic Viewer (P.V 1.15.3) software to quantify positive cells at the single-cell level.
Functional enrichment analysis
To identify genes associated with CD47 expression, RNAseq data from the TCGA database were analyzed. Genes positively correlated with CD47 expression, displaying a —log2(fold change)— ≥ 2 and a corrected P-value <0.05, were selected for further analysis. Enrichment analysis of the 2,486 CD47 co-expressed genes was conducted using the “clusterProfiler” package. To explore the associated biological processes, molecular functions, and cellular localization, we employed the “ggplot2” package for visualization. In addition, KEGG pathway enrichment analysis was performed to elucidate the signaling pathways in which these genes are involved in COAD.
GSEA enrichment
To assess the impact of differential CD47 expression on cell signaling pathways, we analyzed the differentially expressed genes between the CD47 high- and low-expression groups. GSEA was performed using the KEGG.v7.4 dataset with default reference settings. An absolute net enrichment score (NES) >1, false discovery rate (FDR) q < 0.25, and P value < 0.05 were considered to represent significant enrichment.
Expression of CD47 in each cell in the COAD tumor microenvironment (TME)
The TISCH database (http://tisch.comp-genomics.org/) is a comprehensive resource integrating single-cell sequencing data from 27 different types of cancer. Based on MAESTRO v.1.1.0 , all collected datasets were processed uniformly through a standardized workflow including quality control, batch effect removal, cell clustering, differential expression analysis, cell type annotation, malignant cell classification, and GSEA. batch effects were corrected for if the median entropy was below 0.7 using Seurat v.3.1.2 . For each cluster, TISCH used the Wilcoxon test to identify differentially expressed genes (DEGs) based on log-transformed fold change (—logFC—>=0.25) and annotated the cell clusters using the marker-based annotation method employed in MAESTRO. Marker genes for each cell type were collected from published resources . For this study, we utilized the CRC_GSE139555 and CRC_GSE136394 datasets to investigate the expression of CD47 in individual cells within the TME of COAD. The results were visualized using a heatmap to present the expression patterns.
Analysis of tumor immune infiltration and macrophage polarization
We utilized the TIMER database (http://cistrome.shinyapps.io/timer) to perform molecular characterization of tumor-immune interactions. The correlation between CD47 expression levels and the abundance of six distinct tumor-infiltrating immune subpopulations (CD8 + T cells, CD4 + T cells, macrophages, B cells, dendritic cells, and neutrophils) was calculated, P < 0.05 was considered statistically significant.
Correlation analysis of biomarkers and treatment response
The Gene Set Cancer Analysis (GSCALite) (bioinfo.life.hust.edu.cn/web/GSCALite/) server (Liu et al., 2018) was employed to analyze the gene expression profile of CD47 and calculate the area under the dose–response curve (AUC) values across various cancer cell lines. To evaluate the treatment response to CD47 and overall survival (OS) in different cancer types, we compared seven standardized biomarkers using the biomarker assessment module of the TIDE server. These biomarkers included T cell clonality (T.clonality), B cell clonality (B.clonality), TIDE, microsatellite instability (MSI) score, tumor mutation burden (TMB), CD274 (PD-L1), and interferon-gamma (IFNG) (Fu et al., 2020). Their correlations with tumor immune response were analyzed to assess predictive value.
Statistical analysis
All experimental data were analyzed using SPSS 22.0 statistical software. For dichotomous data involving clinicopathological parameters, the chi-square test was applied, with continuity correction when expected counts were less than 5. Quantitative data were compared using the independent sample t-test. Factors found to be significant in univariate analysis were further evaluated by multivariate Cox regression to identify independent risk factors for survival. A P-value of <0.05 was considered statistically significant.
Results
The mRNA expression of the CD47 gene in COAD
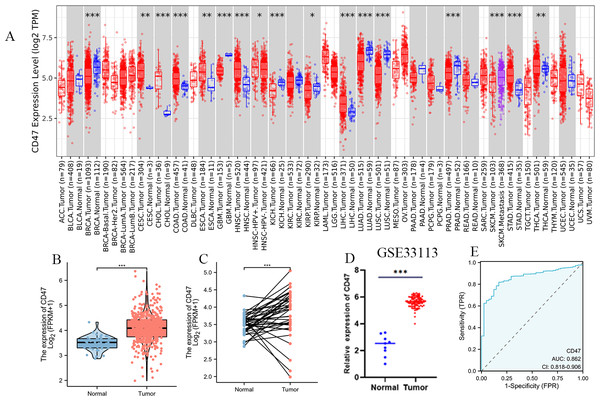
Using the TCGA database, we analyzed CD47 gene expression across various human cancers and their corresponding normal tissues. Our analysis revealed significantly elevated CD47 expression in nine cancer types, including colon cancer, cervical squamous cell carcinoma, bile duct carcinoma, esophageal cancer, head and neck squamous cell carcinoma, LIHC, metastatic cutaneous melanoma, gastric adenocarcinoma, and thyroid cancer (Fig. 1A). In comparisons of unpaired samples, tumor tissues consistently exhibited markedly higher CD47 levels than normal tissues (Fig. 1B). This finding was further confirmed by a paired analysis of 41 normal tissues and their corresponding tumor samples, which demonstrated significantly increased CD47 expression in cancerous tissues (P < 0.001) (Fig. 1C). Additionally, cross-validation with the GEO dataset (GSE33113) corroborated our results, showing heightened CD47 expression in COAD tissues compared to normal tissues (Fig. 1D). Receiver operating characteristic (ROC) curve analysis highlighted CD47’s strong diagnostic potential, with high sensitivity (0.818) and specificity (0.829) for predicting patient outcomes (AUC = 0.862) (Fig. 1E).
Figure 1: Expression of CD47 in different types of tumors and COAD.
(A) Expression of CD47 in different types of tumors compared with normal tissues in TCGA and GTEx databases; (B) higher expression of CD47 in COAD than in the mismatched normal tissue; (C) CD47 is higher in COAD thanin the matched normal tissue (D) GEO databaseverifies the expression of CD47in COAD; (E) receiver operating characteristic curve of CD47 in the TCGA database.Expression of CD47 in tissue samples of COAD patients
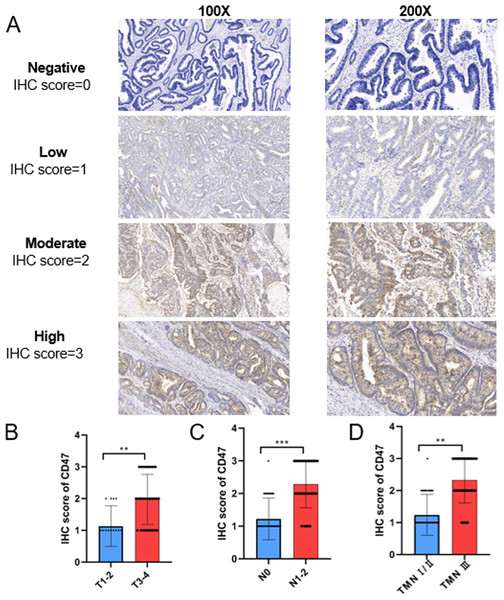
CD47 protein expression was evaluated in 90 COAD tissue samples. The results showed that CD47 was predominantly localized to the cell membrane and/or cytoplasm of tumor cells, with a positivity rate of 91.11% (80/90). Figure 2A presents the variation in CD47 staining intensity as assessed by IHC. Statistical analysis revealed significantly higher CD47 IHC scores in T3-4 tumors compared to T1-2 tumors (Fig. 2B). Additionally, CD47 expression was significantly elevated in N1/2 COAD patients compared to N0 patients (Fig. 2C). Furthermore, patients with TNM stage III/IV disease exhibited significantly higher CD47 expression than those with TNM stage I/II disease (Fig. 2D).
Figure 2: Representative images and statistical analysis of IHC for CD47 in COAD tissues.
(A) Representative images of IHC staining of CD47 with negative, weak, moderate, and strong expression. (B) Statistical analysis of CD47 expression in T1/2 and T3/4 stage COAD samples based on IHC scores. (C) The expression of CD47 in patients with lymph node metastasis was significantly higher than that in patients without lymph node metastasis. (D) The expression of CD47 in TNM stage III/IV patients was significantly higher than that in TNM stage I/II patients.Correlation between CD47 expression and the clinical characteristics of COAD patients
In this study, high CD47 expression was defined by an IHC score of 3 or higher. We examined the relationship between CD47 expression levels in tumor tissues and various clinicopathological factors, including gender, age at diagnosis, and tumor stage (T, N, and overall clinical stage). CD47 expression was significantly associated with both the N stage (P < 0.001) and the overall clinical stage of COAD (P < 0.001). However, no significant correlation was observed between CD47 expression and gender, age, or the T stage, as shown in Table 1.
| Characteristic | Low expression of CD47 | High expression of CD47 | p |
|---|---|---|---|
| n | 69 | 21 | |
| T stage, n(%) | 0.139 | ||
| T1 | 3 (3.3%) | 0 (0%) | |
| T2 | 11 (12.2%) | 0 (0%) | |
| T3 | 47 (52.2%) | 17 (18.9%) | |
| T4 | 8 (8.9%) | 4 (4.4%) | |
| N stage, n (%) | <0.001 | ||
| N0 | 45 (50%) | 1 (1.1%) | |
| N1 | 12 (13.3%) | 10 (11.1%) | |
| N2 | 12 (13.3%) | 10 (11.1%) | |
| Clinical stage, n (%) | <0.001 | ||
| I | 7 (7.8%) | 0 (0%) | |
| II | 38 (42.2%) | 1 (1.1%) | |
| III | 24 (26.7%) | 20 (22.2%) | |
| Age, n (%) | 0.744 | ||
| <60 | 11 (12.2%) | 4 (4.4%) | |
| ≥60 | 58 (64.4%) | 17 (18.9%) | |
| Gender, n (%) | 1.000 | ||
| Female | 38 (42.2%) | 11 (12.2%) | |
| Male | 31 (34.4%) | 10 (11.1%) |
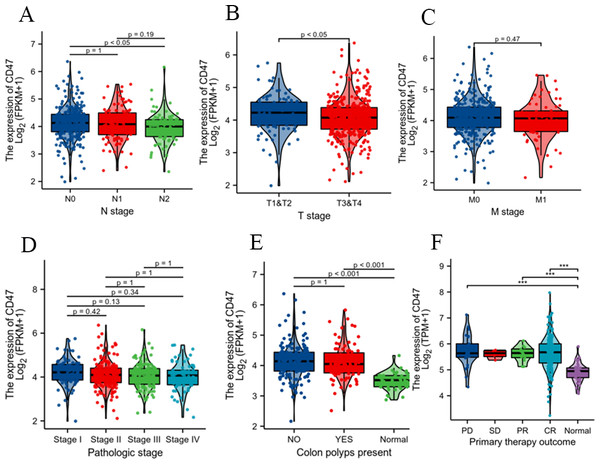
To further validate our findings, we analyzed clinicopathological data from COAD patients in the TCGA database. This analysis confirmed the positive correlation between CD47 expression and the N stage (Fig. 3A) and T stage (Fig. 3B). However, no significant association was observed with the M stage (Fig. 3C), pathological stage (Fig. 3D), or the presence of colon polyps (Fig. 3E). Additionally, CD47 expression was significantly higher in patients with disease progression (PD), partial response (PR), and complete response (CR) compared to normal tissues. In contrast, there was no significant difference in CD47 expression between the stable disease (SD) group and normal tissues (Fig. 3F).
Figure 3: Associations between CD47 expression and clinicopathological characteristics.
Data are shown for (A) N stage; (B) T stage; (C) M stage; (D) pathologic stage; (E) colonpolyps present; (F) primary therapy outcome.Analysis of the independent prognosis
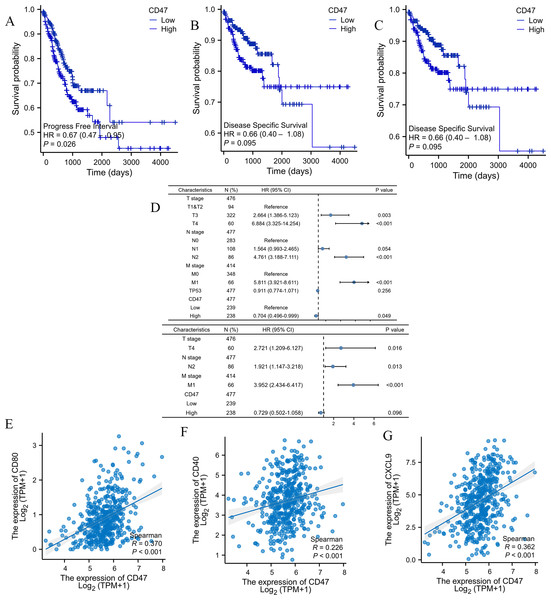
The relationship between CD47 expression and the prognosis of COAD patients was evaluated using the Kaplan–Meier method. Notably, high CD47 expression was associated with longer progression-free survival (PFI) compared to low CD47 expression. However, no significant differences were observed in overall survival (OS) or disease-specific survival (DSS) (PFI: HR = 0.57, 95% CI [0.30–1.11], P = 0.025; OS: HR = 0.82, 95% CI [0.37–1.81], P = 0.144; DSS: HR = 0.9, 95% CI [0.35–2.29], P = 0.093) (Figs. 4A, 4B, 4C).
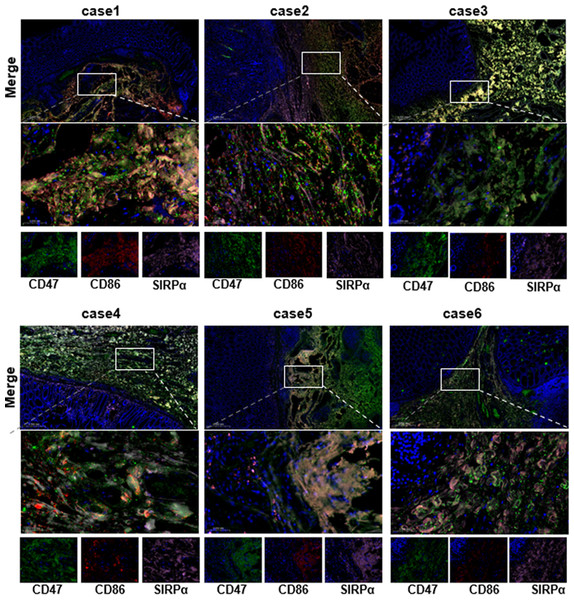
Figure 4: Co-expression of CD47, SIPα, and M1 macrophage surface marker CD86 in COAD was analyzed by multiplex immunofluorescence staining.
To further examine the impact of clinicopathological features on patient outcomes, we performed univariate and multivariate independent prognostic analyses. Univariate analysis identified CD47 expression and TNM stage as significant prognostic factors (P < 0.05), while TP53 was not significantly associated with prognosis (P = 0.256). Interestingly, multivariate analysis demonstrated that high CD47 expression had an HR below 1.0, indicating a favorable prognosis (Fig. 4D and Table 2), which contrasts with our earlier findings. Given the observed correlation between high CD47 expression and advanced TNM stage, it is perplexing that high CD47 expression would be linked to longer PFI in COAD patients. To investigate this discrepancy, we explored the immunophenotypic and functional differences between M1 and M2 tumor-associated macrophages (TAMs), focusing on the relationship between CD47 expression and M1 macrophage infiltration. Our analysis revealed a positive correlation between CD47 expression and the M1 macrophage markers CD86 (Spearman’s R = 0.356, P < 0.001), CXCL9 (Spearman’s R = 0.36, P < 0.001), and CD80 (Spearman’s R = 0.368, P < 0.001) (Figs. 4E–4G).
| Characteristics | Total (N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| T stage | 476 | ||||
| T1&T2 | 94 | Reference | |||
| T3 | 322 | 2.664 (1.386–5.123) | 0.003 | 1.540 (0.774–3.065) | 0.218 |
| T4 | 60 | 6.884 (3.325–14.254) | <0.001 | 2.721 (1.209–6.127) | 0.016 |
| N stage | 477 | ||||
| N0 | 283 | Reference | |||
| N1 | 108 | 1.564 (0.993–2.465) | 0.054 | 0.856 (0.505–1.452) | 0.565 |
| N2 | 86 | 4.761 (3.188–7.111) | <0.001 | 1.921 (1.147–3.218) | 0.013 |
| M stage | 414 | ||||
| M0 | 348 | Reference | |||
| M1 | 66 | 5.811 (3.921–8.611) | <0.001 | 3.952 (2.434–6.417) | <0.001 |
| TP53 | 477 | 0.911 (0.774–1.071) | 0.256 | ||
| CD47 | 477 | ||||
| Low | 239 | Reference | |||
| High | 238 | 0.704 (0.496–0.999) | 0.049 | 0.729 (0.502–1.058) | 0.096 |
Based on these findings, we speculate that the prognostic significance of CD47 may be diminished in COAD patients with increased infiltration of M1 polarized macrophages. However, further studies are needed to clarify CD47’s role in COAD and to better understand its correlation with the heterogeneity of the tumor immune microenvironment.
Immune infiltration analyses in COAD
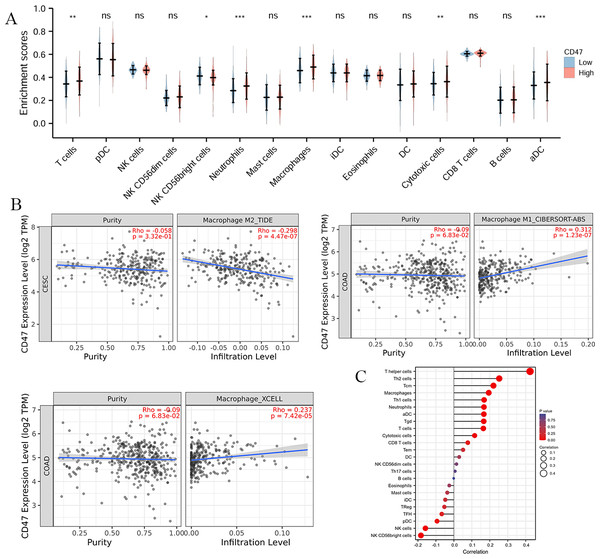
The OS rates in COAD were strongly influenced by tumor immune infiltration. Analysis of 24 different immune cell subtypes showed significant increases in T cells (P = 0.001), neutrophils (P < 0.001), macrophages (P < 0.001), cytotoxic cells (P = 0.006), and activated dendritic cells (aDCs) (P < 0.001) in the high CD47 expression group (Fig. 5A).
Figure 5: Prognostic values of CD47 expression in patients with COAD evaluated by the Kaplan–Meier method.
(A) Overall survival; (B) disease-specific survival; (C) progress-free interval. (D) Forest map based on multivariate Cox analysis for Disease Free Interval; (E–G) correlation analysis of CD47 and surface markers in M1 macrophages. HR, hazard ratio; CI, confidence interval.The CD47-SIRPα signaling pathway has been reported to negatively regulate macrophage phagocytosis. SIRPα expression on TAMs inhibits intracellular FcγR signaling, resulting in reduced phagocytic activity and allowing tumor cells to evade immune detection (Murata et al., 2014; McCracken, Cha & Weissman, 2015; Gül & Van Egmond, 2015). To explore this further, we examined the correlation between CD47 expression and macrophage infiltration levels. Our analysis revealed a positive correlation between CD47 expression and M1 macrophages (r = 0.312, P < 0.001), as well as total macrophage infiltration (r = 0.237, P < 0.001). In contrast, a negative correlation was observed between CD47 expression and M2 macrophages (r = −0.298, P < 0.001) (Fig. 5B). Additionally, CD47 expression showed a significant negative association with NK CD56bright cells (r = 0.170, P < 0.001) and plasmacytoid dendritic cells (pDCs) (r = 0.119, P < 0.001) (Fig. 5C).
Multiplex immunofluorescence analysis
To gain a deeper understanding of CD47’s role in COAD, we performed multiplex immunofluorescence staining to assess the co-expression of CD47 with M1 macrophage marker CD86 and SIRPα in COAD tissues. Six COAD specimens, including adjacent non-tumor tissues, were analyzed to visually assess CD47 expression. Immunofluorescence labeling was conducted with CD86 (red), CD47 (green), SIRPα (pink), and DAPI (blue). The results showed that CD47, SIRPα, and CD86 were co-expressed in COAD tissues, with significantly higher expression levels of CD47 and SIRPα compared to CD86 (Fig. 6). Further confirmation of intercellular interactions within the COAD TME was obtained through multiplex IHC, which revealed that CD47 exerts immunosuppressive effects primarily via the CD47/SIRP α axis. These findings suggest that targeting CD47 with specific blockers may enhance macrophage-mediated phagocytosis of tumor cells, thereby inhibiting tumor progression in COAD (Gu et al., 2018).
Figure 6: Association between CD47 expression and immune infiltration in COAD.
(A) Expansion levels of immune cells of 24 subtypes in the high and low CD47 expression groups. (B) Correlation between CD47 expression and macrophages. (C) Correlation between CD47 expression and the level of immune inflation.Correlation between CD47 and the tumor immune microenvironment heterogeneity
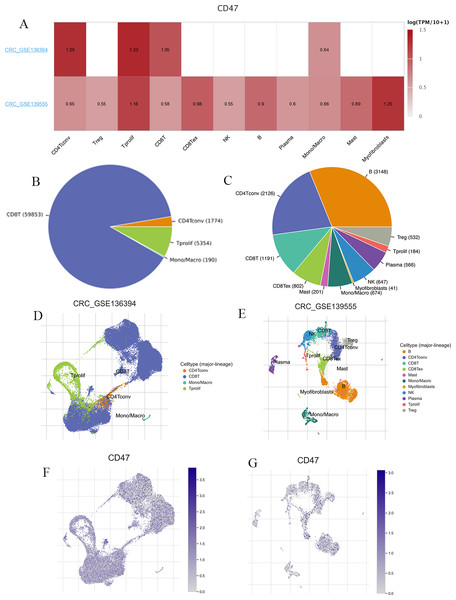
We utilized two datasets from the TISCH database, CRC_GSE139555 and CRC_GSE136394, to examine CD47 expression in immune cells within the TME (Fig. 7A). The CRC_GSE139555 dataset contains data from primary CRC patients, while the CRC_GSE136394 dataset focuses primarily on metastatic tumors (Wu et al., 2020; Lu et al., 2019). In the metastatic CRC_GSE136394 dataset, only four cell types were identified: CD4Tconv, CD8T, Mono/Macro cells, and Tprolif, with CD8T cells being the most abundant (Fig. 7B). In contrast, the CRC_GSE139555 dataset revealed 11 distinct cell types (Fig. 7C). Figures 7D–7G, visually present the distribution of these immune cells, highlighting significant differences in cellular composition and distribution between primary and metastatic CRC. The expression levels of CD47 were found to vary across different cell types, and these cellular components differed between primary and metastatic patients. This variation may explain the heterogeneity observed within the CRC TME.
Figure 7: CD47-related cell type distribution using scRNA seq database.
(A) Expression levels of CD47; (B3E) the cell types and their distribution in CRC_GSE139555 and CRC_GSE136394. (F–G) Distribution of CD47 in different cells in CRC_GSE139555 and CRC_GSE136394 datasets.CD47 participates in tumor immune evasion through different T-cell rejection
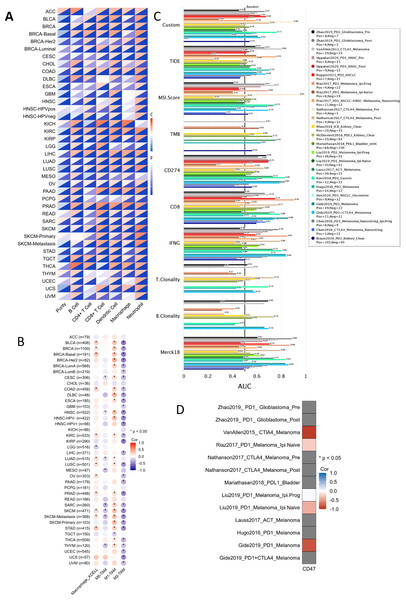
Among the 22 TCGA cancer types and subtypes assessed for tumor immune infiltration, a significant positive correlation was observed between CD47 expression and immune cell infiltration in three cancer types: COAD, kidney renal clear cell carcinoma (KIRC), and LIHC (Fig. 8A). Specifically, LIHC showed a strong correlation with immune cells (r = 0.28–0.47, P < 0.05), renal clear cell carcinoma exhibited a correlation of (r = 0.26–0.45, P < 0.05), and colorectal adenocarcinoma demonstrated a correlation of (r = 0.17–0.42, P < 0.05). These findings suggest a strong association between CD47 expression and immune cell infiltration in these cancers. Additionally, we analyzed the correlation between CD47 expression and four key immunosuppressive cell types known to promote T-cell rejection: myeloid-derived suppressor cells (MDSCs), cancer-associated fibroblasts (CAFs), regulatory T cells (Tregs), and M2 tumor-associated macrophages (M2-TAMs). CD47 expression was significantly correlated with these immunosuppressive cells in most cancer types, with the exception of adrenocortical carcinoma (ACC), cholangiocarcinoma (CHOL), renal chromophobe (KICH), pheochromocytoma and paraganglioma (PCPG), and rectal adenocarcinoma (PEAD). Notably, CD47 was positively correlated with MDSC and Treg infiltration, while showing a negative correlation with M2-TAM infiltration (Fig. 8B).
Figure 8: Correlation between CD47 expression and immune detection points.
(A) CD47 with infiltration of six immune cell types; (B) CD47 and macrophage infiltration; (C) bar plot showing correlation between CD47 and standardized cancer-immune evasion markers in immune checkpoint (ICB), area under the receiver operating characteristic curve (AUC) to evaluate the predictive performance of test biomarker response to ICB status; (D) heat map showing CRISPR screening between CD47 and lymphocyte-mediated tumor killing in the ICB sub-cohort.These results suggest that CD47 plays a role in T-cell rejection through multiple mechanisms. Furthermore, we compared CD47 with standardized biomarkers in terms of response prediction and OS biomarker correlations. Using 15 feature curves with an AUC greater than 0.5, we found that CD47 had a higher predictive value than tumor mutation burden (TMB) and T-cell clonality in seven and six immunotherapy subpopulations, respectively. Additionally, CD47 exhibited promising treatment outcomes in melanoma when combined with CTLA-4 inhibitor therapy (ICB_VanAllen2015) and PD-1 inhibitor therapy (ICB_Gide2019_PD1) (Fig. 8D).
Immune-related pathways regulated by CD47 in COAD
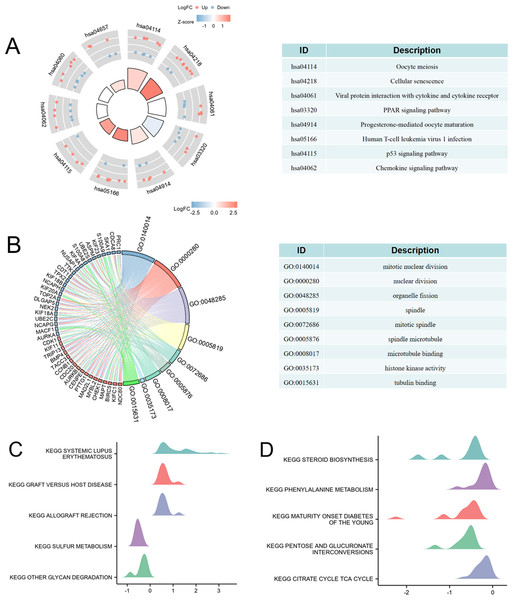
To explore the relationship between high CD47 expression and immune regulation, we performed KEGG, GO, and GSEA analyses. Initially, differential expression analysis identified genes associated with high CD47 expression in COAD (Fig. S1). KEGG pathway enrichment was then conducted to investigate the biological processes linked to elevated CD47 expression. The enriched KEGG pathways were primarily associated with oocyte meiosis, cellular senescence, viral protein interactions with cytokine and cytokine receptors, PPAR signaling, progesterone-mediated oocyte maturation, human T-cell leukemia virus 1 infection, p53 signaling, and chemokine signaling pathways (Fig. 9A). These results highlight a strong correlation between high CD47 expression and immune cell infiltration, as well as regulation of the immune microenvironment in COAD.
Figure 9: CD47-related differentially expressed genes (DEGs) and functional enrichment analysis of CD47 in Colon adenocarcinoma using KEGG, GO and GSEA.
(A) KEGG analysis of DEGs. (B) GO analysis of DEGs. (C–D) GSEA analysis of DEGs. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes.GO enrichment analysis revealed that the differentially expressed genes were involved in nine key biological processes: mitotic nuclear division, nuclear division, organelle fission, spindle formation, mitotic spindle organization, spindle microtubule assembly, microtubule binding, histone kinase activity, and tubulin binding (Fig. 9B). Furthermore, GSEA analysis showed that high CD47 expression was significantly associated with processes such as systemic lupus erythematosus (NES = 1.57, P = 0.00), graft-versus-host disease (NES = 1.60, P = 0.00), allograft rejection (NES = 1.53, P = 0.04), and several metabolic pathways, including sulfur metabolism (NES = −2.248, p = 0.00), glycan degradation (NES = −1.84, P = 0.00), steroid biosynthesis (NES = −1.93, P = 0.00), phenylalanine metabolism (NES = −2.11, p = 0.00), maturity-onset diabetes of the young (NES = −2.53, P = 0.01), pentose and glucuronate interconversions (NES = −1.87, P = 0.02), and the citrate (TCA) cycle (NES = −1.71, P = 0.02) (Figs. 9C–9D).
Discussion
Colon cancer is one of the most common malignancies of the digestive tract worldwide, with a high recurrence rate and poor prognosis (Ahiko et al., 2021; Dai et al., 2020; Biondo et al., 2019) . Therefore, identifying novel diagnostic and therapeutic targets is of paramount importance. In this study, we used the TCGA online database to examine the aberrant expression of CD47 across 17 tumor types. Notably, we found elevated CD47 expression in COAD, cervical cancer, bile duct cancer, esophageal cancer, and hepatocellular carcinoma, while lower expression was observed in glioblastoma multiforme, renal papillary cell carcinoma, and renal clear cell carcinoma. These findings align with previous research reporting CD47 overexpression in various tumors, including colon cancer (Gong et al., 2023), hepatocellular carcinoma (Koga et al., 2021), acute lymphoblastic leukemia (Morse et al., 2023), cervical cancer (Yordanov et al., 2022), breast cancer (Chen et al., 2023), and gastric cancer (You et al., 2022).
In our study, we examined CD47 expression in COAD by analyzing tissue samples from 90 patients who had not undergone chemoradiotherapy. These samples were subjected to immunohistochemical analysis, revealing a significant increase in CD47 expression in COAD tissues. We also found that elevated CD47 levels were associated with unfavorable clinicopathological features, particularly advanced T (T3 and T4) and N stages. Furthermore, our data suggest that increased CD47 expression may serve as an independent marker for predicting reduced disease-free intervals (DFI) in colon cancer patients. Given the critical role of tumor immunity in clinical research, it is noteworthy that some studies (Duan et al., 2023) have shown that CD47 can attenuate the anti-tumor immune response, especially in Epstein-Barr virus-associated gastric cancer, by modulating T cell infiltration. This suggests that CD47 could be a promising immunotherapeutic target in this subtype of gastric cancer. Additionally, other research (Sugimura-Nagata et al., 2021) on colon cancer has highlighted the potential of targeting the CD47-SIRPα pathway as a therapeutic strategy for CRC. However, our study primarily focused on the prognostic implications of CD47 and its association with immune factors in COAD, rather than exploring the detailed molecular mechanisms underlying CD47 regulation.
The TME of COAD is widely recognized for its marked heterogeneity (Wijnands et al., 2021; Mimori et al., 2018). To gain insights into how CD47 influences the TME, we utilized the TISCH single-cell database. By examining primary and metastatic CRC, we identified distinctive immune cell distributions at the tumor site. Primary COAD tumors exhibited a greater variety of cell types, including B cells, mast cells, CD8 Tex, myofibroblasts, NK cells, and Trges. Regarding CD47 expression, the CRC_GSE136394 dataset displayed higher levels in CD4Tconv, T-proliferating, and CD8 T cells than the CRC_GSE139555 dataset. These observations indicate that the TME of primary CRC may differ from that of metastatic CRC, contributing to the overall heterogeneity of this disease.
The dynamic interplay between tumor molecular characteristics and the immune microenvironment significantly influences tumor development, leading to varying clinical outcomes. Notably, tumor-infiltrating immune cells are known to contribute to an impaired or dysfunctional T-cell phenotype (Joyce & Fearon, 2015), fostering conditions conducive to tumor evasion, progression, metastasis, and resistance to treatment. By analyzing the TCGA database, our research probed the association between CD47 expression and tumor immune cell infiltration. We identified a significant correlation of CD47 with immune cell infiltration in KIRC, COAD, and LIHC. These insights underscore the potential role of CD47 in promoting tumor immune escape by fostering a dysfunctional T-cell environment.
CD47’s influence extends across various cancer-related signaling pathways. Notably, the CD47/SIRPα axis is a key mediator in cancer immune evasion and immunotherapy strategies (Jia et al., 2021). Additionally, in a study on murine hepatocellular carcinoma, blocking CD47 was shown to bolster antitumor immunity via the CD103(+) dendritic cell-NK cell axis (Wang et al., 2022). However, these insights do not fully unravel the workings of CD47 in COAD, highlighting the need for further exploration of its biological roles and related signaling pathways in this context. In our research, KEGG analysis was employed. This revealed that high CD47 expression significantly correlated with several pathways, including oocyte meiosis, cellular senescence, viral protein interactions with cytokines and cytokine receptors, the PPAR signaling pathway, progesterone-mediated oocyte maturation, human T-cell leukemia virus 1 infection, the p53 signaling pathway, and chemokine signaling. The PPAR, part of the nuclear receptor transcription factor superfamily, is notably activated via ligand-binding and plays a crucial role in regulating adipocyte differentiation and fatty acid metabolism (Faghfouri et al., 2021). Recent studies have also highlighted PPAR signaling’s pivotal role in modulating T cell and macrophage functions and its influence on the quantity and immune activity of regulatory T cells in visceral adipose tissue (Sica et al., 2015). Moreover, PPAR signaling is implicated in the differentiation of macrophages towards pro-inflammatory or anti-inflammatory states (Liu et al., 2020). These findings suggest that CD47 might regulate macrophage or regulatory T cell activity in COAD through the PPAR signaling pathway.
An interesting result that emerged in this study was that CD47 was highly expressed in COAD, but the time to PFI was prolonged in COAD patients with high CD47 expression. On the one hand, it has been shown that CD47 expression in tumor cells correlates with poor prognosis. For example, in patients with acute myeloid leukemia (AML), the positive rate of CD47 expression was significantly higher than that in patients with benign hematological disorders, and the complete remission rate was lower in CD47-positive AML patients, which suggests that high expression of CD47 may be associated with a poorer prognosis in AML patients (Majeti et al., 2009). In addition, high expression of CD47 in lymphomas has been found to be associated with poor prognosis, especially in classic Hodgkin’s lymphoma and Sezary syndrome (Johnson et al., 2019). However, on the other hand, some studies have not clearly indicated a direct association between CD47 expression and prognosis, or have found the opposite result. For example, in studies of non-small cell lung cancer (NSCLC), CD47 has been identified as an inverse prognostic factor in NSCLC, but anti-cd47-based combination therapy remains a promising approach for EGFR-mutant NSCLC (Hu et al., 2024a; Hu et al., 2024b).
This suggests that findings on the relationship between CD47 expression and prognosis are inconsistent and possibly contradictory.The function of CD47 is complex, and there is still a long way to go to study it.
In the present study, high CD47 expression was positively correlated with the level of M1 macrophage infiltration, which may be an important factor influencing the prognosis of COAD. The presence of M1-type macrophages in COAD has been well documented, and they may affect various aspects of tumor progression, including proliferation, migration, apoptosis, and the cell cycle of tumor cells, etc. After the conditioning of colorectal cancer cells by the medium-conditioned action of M1 macrophages, M1-type macrophages induced the activation of caspase pathway in colorectal cancer cells (Lee et al., 2020). Recently, however, it has also been suggested that M1-type macrophages accumulate at the invasion front of early colorectal cancer, and that they may upregulate SAA1 through the IL1β signaling pathway, which promotes the migration and invasion of COAD cells (Sudo et al., 2021). It has also been suggested that the inflammatory environment created by M1-type macrophages promotes colorectal cell EMT (Marcuello et al., 2018). How M1-type macrophages play a role in the different TMEs of colorectal cancer and the specific mechanisms that influence the growth and shepherding of colorectal cancer need further investigation. There are relatively few basic studies and clinical trials on the use of M1-type macrophages as an immunotherapeutic target, and further studies are still needed. In addition, CD47 protein is expressed to different degrees in both stromal cells and tumor cells. Therefore, the prognostic value of CD47 expression in different cell types should be further distinguished in the future.
Overall, the current findings on the relationship between CD47 expression and prognosis are contradictory, which may be due to differences in study methods and samples, as well as the specificity of the study subjects. Future studies need to further explore the specific mechanisms of CD47’s role in different types of cancer and how to improve patients’ prognosis by regulating CD47 expression.
Conclusion
In conclusion, our study provides evidence that elevated levels of CD47 expression have an impact on the TME in COAD. Specifically, heightened CD47 expression may promote the M1 phenotype of tumor-associated macrophages, enabling them to exhibit enhanced antigen-presenting capacity. Conversely, tumor cells employ highly expressed CD47 to inhibit macrophage phagocytosis by binding to SIRPα receptors on their surface, leading to immune evasion. Therefore, our findings have significant implications for understanding the role of CD47 in the TME and its influence on chemotherapy response. These insights hold promising clinical significance, as they may contribute to improving therapeutic efficacy and guiding treatment decisions.
This study has several limitations. First, the number of patients that could be examined was not large enough, and sufficient clinical information was not obtained, such as the failure to differentiate between high microsatellite unstable colon cancers and higher microsatellite stable colorectal cancers. Therefore, prognostic analysis of tumor mutational load associated with CD47 expression could not be performed. A second limitation was that the assessment of CD47-positive and inflammatory cells in the tumor microenvironment was not addressed. Overall, it was possible to differentiate between tumor and non-tumor cells and to identify the spectrum of inflammatory cells based on morphological findings. However, a more detailed assessment was not performed as IHC was not used. Further understanding should be established in the future by using IHC and other molecular techniques to elucidate the properties of the tumor microenvironment, possibly. The third limitation is that cellular experiments were not performed to validate the function of CD47 in COAD, which could be further validated in the future by constructing CD47-interfering or overexpressing cell lines using gene editing techniques.
Supplemental Information
CD47 high expression associated with DEGs in colon adenocarcinoma.(A) the TCGAdatabase; (B) the GEO database
(A) the TCGA database; (B) the GEO database.