Identification and expression analysis of EDR1-like genes in tobacco (Nicotiana tabacum) in response to Golovinomyces orontii
- Published
- Accepted
- Received
- Academic Editor
- Ivo Feussner
- Subject Areas
- Agricultural Science, Genetics, Genomics, Plant Science
- Keywords
- Powdery mildew, ENHANCED DISEASE RESISTANCE1, Tobacco, Phylogenetic analysis, Expression patterns
- Copyright
- © 2018 Wu et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2018. Identification and expression analysis of EDR1-like genes in tobacco (Nicotiana tabacum) in response to Golovinomyces orontii. PeerJ 6:e5244 https://doi.org/10.7717/peerj.5244
Abstract
ENHANCED DISEASE RESISTANCE1 (EDR1) encodes a Raf-like mitogen-activated protein kinase, and it acts as a negative regulator of disease resistance and ethylene-induced senescence. Mutations in the EDR1 gene can enhance resistance to powdery mildew both in monocotyledonous and dicotyledonous plants. However, little is known about EDR1-like gene members from a genome-wide perspective in plants. In this study, the tobacco (Nicotiana tabacum) EDR1-like gene family was first systematically analyzed. We identified 19 EDR1-like genes in tobacco, and compared them to those from Arabidopsis, tomato and rice. Phylogenetic analyses divided the EDR1-like gene family into six clades, among them monocot and dicot plants were respectively divided into two sub-clades. NtEDR1-1A and NtEDR1-1B were classified into clade I in which the other members have been reported to negatively regulate plant resistance to powdery mildew. The expression patterns of tobacco EDR1-like genes were analyzed after plants were challenged by Golovinomyces orontii, and showed that several other EDR1-like genes were induced after infection, as well as NtEDR1-1A and NtEDR1-1B. Expression analysis showed that NtEDR1-13 and NtEDR1-16 had exclusively abundant expression patterns in roots and leaves, respectively, and the remaining NtEDR1-like members were actively expressed in most of the tissue/organ samples investigated. Our findings will contribute to further study of the physiological functions of EDR1-like genes in tobacco.
Introduction
Powdery mildew, an important fungal disease in agriculture and horticulture, is caused by ascomycetes of the order Erysiphales (Dean et al., 2012; Glawe, 2008). Worldwide, powdery mildews colonize a wide variety of plant species—over 650 monocots and over 9,000 dicots (Schulzelefert & Vogel, 2000). They cause significant harvest losses in crops such as wheat, barley and tomato (Dean et al., 2012), ornamental plants such as roses (Horst, Kawamoto & Porter, 1992; Linde et al., 2006; Palmer & Henneberry, 1960) and fruits like grapevine (Donald et al., 2002). The current universal method for controlling powdery mildew is by application of fungicide, but this can have a serious environmental impact. Research has detected high concentrations of residual fungicides in food and crops. These chemicals can reach human cells via the food chain and negatively affect cellular metabolism (Pirozzi et al., 2016; Yang et al., 2018). Thus, identifying genes with functions in fundamental plant defense may have potential for reducing production losses caused by powdery mildew. In 1998, Frye and Innes found an Arabidopsis mutant that displayed ENHANCED DISEASE RESISTANCE1 (EDR1) to Pseudomonas syringae and Golovinomyces cichoracearum (formerly named Erysiphe cichoracearum) (Frye & Innes, 1998). Functional studies of EDR1 genes have been reported in Arabidopsis, wheat, rice and tomato (Frye & Innes, 1998; Gao et al., 2015; Shen et al., 2011; Zhang et al., 2017).
In Arabidopsis, AtEDR1 encodes Raf-like mitogen-activated protein kinase. Loss-of-function mutants of EDR1 are resistant to the biotrophic pathogens G. cichoracearum (Frye & Innes, 1998) and Hyaloperonospora arabidopsidis (van Hulten et al., 2006) and also show enhanced susceptibility to hemibiotrophic Colletotrichum higginsianum and necrotrophic Alternaria brassicicola (Hiruma et al., 2011). Thus, EDR1 mutants can obtain broad-spectrum resistance. In wheat, knockdown TaEDR1 mutants from VIGS or RNAi showed increased resistance to virulent isolates of Blumeria graminis f. sp. tritici (Zhang et al., 2017). Additionally, wheat EDR1 plants generated by simultaneous modification of the three homologs of TaEDR1 with CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated) technology, did not exhibit powdery mildew-induced cell death (Zhang et al., 2017). The sequence ortholog of Arabidopsis EDR1 in rice, OsEDR1, has a negative function in the defense response (Shen et al., 2011). The expression of OsEDR1 was induced by wounding, jasmonic acid, salicylic acid, ethylene, abscisic acid, hydrogen peroxide, fungal elicitor chitosan, drought, high salt, sugar and heavy metals (Kim et al., 2003). The RNAi plants or T-DNA insertion mutants of OsEDR1 had enhanced resistance to bacterial pathogen Xanthomonas oryzae pv. oryzae (Shen et al., 2011). However, silencing of two EDR1 homologs (Solyc01 g097980 and Solyc06 g068980) separately did not confer significant resistance against Oidium neolycopersici in tomato (Gao et al., 2015). Whether simultaneous knockdown of these two homologs could enhance powdery mildew resistance needs further investigation.
The EDR1 protein consists of an N-terminal domain of unknown function and a C-terminal kinase domain (Tang & Innes, 2002). The C-terminal of EDR1 encodes a serine/threonine-protein kinase with homology to CONSTITUTIVE TRIPLE RESPONSE1 (Tang, Christiansen & Innes, 2005), a negative regulator of ethylene responses (Cao et al., 1997; Kieber et al., 1993). The N-terminal regulatory domain of EDR1 could interact with MKK4 and MKK5 to negatively regulate the MKK4/MKK5–MPK3/MPK6 kinase cascade pathway (Zhao et al., 2014). It was also reported that EDR1 could act as a suppressor of disease resistance and programmed cell death (for both abiotic and biotic stresses) by adjusting signal processing through the hormone-mediated pathways of salicylic acid, ethylene and abscisic acid (Frye & Innes, 1998; Tang, Christiansen & Innes, 2005), and as a positive regulator of expression of plant defensins (Hiruma et al., 2011).
Although the EDR1 pathway is highly conserved in crop plants (Frye, Tang & Innes, 2001), whether one EDR1 plays a main role or several EDR1 homologs work together to function as negative regulators of plant defense needs further detailed analysis for a specific species. For example, in tomato, RNAi of the two EDR1 homologs separately did not produce significant resistance to O. neolycopersici (Gao et al., 2015). It also revealed that knockout of three EDR1 homologs in wheat did not produce complete resistance (Zhang et al., 2017). Thus, complete and systematic study of the EDR1-like genes is needed and will help to find potential target genes for plant breeding. This study involved the first systematic identification and analysis of EDR1-like genes in plants. We aim to provide useful information for further exploring the physiological function of EDR1-like genes in tobacco and other plant species.
Materials and Methods
Identification of the EDR1-like gene family
The genome, gene and protein sequences of Arabidopsis, tomato and rice were downloaded from PlantGDB (http://www.plantgdb.org/), and those of tobacco from the Sol Genomics Network (http://solgenomics.net/organism/). To identify EDR1-like genes in the above-mentioned species, two EDR1 proteins from Arabidopsis (GenBank accession: ABR45974.1) and wheat (GenBank accession: AAU89661.2) were used as the query subjects in a reciprocal Basic Local Alignment Search Tool Protein (BLASTP) analysis with e-values < 1E-50. Then, the online software SMART (http://smart.embl-heidelberg.de/) was used to identify the predicted EDR1-like protein domains. Conserved EDR1 domains and kinase domains within the acquired EDR1 sequences were confirmed by searching NCBI’s conserved domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd/docs/cdd_search.html).
Phylogenetic analysis and conserved domain detection
Phylogenetic trees were constructed in MEGA Version 7.0 (Kumar, Stecher & Tamura, 2016), using the Neighbor-Joining (NJ) method (Saitou, 1987) with parameters of pairwise gap deletion and 1,000 bootstraps. Multiple sequence alignment of the amino acid sequences of EDR1-like proteins was performed using the ClustalX program (version 1.83) (Thompson et al., 1997) and GeneDoc. Maximization for Motif Elicitation program (MEME, http://alternate.meme-suite.org) (Bailey et al., 2006) was used to predict the conserved motif of EDR1-like proteins in N. tabacum.
Gene structure and bioinformatic analysis of tobacco EDR1-like genes
The genomic structure of the EDR1-like gene family was analyzed using GSDS 2.0 (http://gsds.cbi.pku.edu.cn/) (Hu et al., 2015). Protparam (http://web.expasy.org/protparam/) was used to analyze the basic physical and chemical properties of tobacco EDR1-like genes. Subcellular localization of the EDR1-like genes was determined using online software Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) (Chou & Shen, 2008, 2010a, 2010b). The nuclear localization signals (NLS) sequences were predicted by cNLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi#opennewwindow) (Kosugi et al., 2009a, 2009b). Transmembrane helices were predicted using OCTOPUS (http://octopus.cbr.su.se/index.php#opennewwindow) (Viklund & Elofsson, 2008). The three-dimensional (3D) structures were predicted using the online I-TASSER program (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) (Zhang, 2008). The confidence score (C-score) was used to estimate the model’s global accuracy in I-TASSER (Yang & Zhang, 2015), and high C-score indicates a high-quality structure prediction (Roy, Kucukural & Zhang, 2010). The model with highest C-score in all five prediction models was further used to construct 3D model of the target genes. The constructed 3D model was examined and visualized using Chimera 1.2 (https://www.cgl.ucsf.edu/chimera/).
Plant materials and stress treatments
Common tobacco (Nicotiana tabacum cv HHDJY) seeds were surface-sterilized by 10% NaClO, and sown on sterilized plates with MS solid culture medium (PhytoTechnology Laboratories®, Kansas, MO, USA) in a greenhouse maintained at 25 °C and with aday/night cycle of 16/8 h.
For powdery mildew infection, a strain of G. orontii was maintained on 3-month-old tobacco in a greenhouse. Freshly sporulating leaves of heavily infected tobacco were washed in sterile double distilled water (ddH2O), and the collected conidiospores were used immediately. Plants were inoculated by spraying with the inoculum suspension (about 4 × 104 to 5 × 104 spores ml−1). The 3-month-old tobacco leaves were inoculated with G. orontii to give approximately 20–25 spores per cm2 and sampled after 0, 1, 2, 12 and 24 hours post inoculation (hpi). Samples collected at 0 h were used as controls. For tissue/organ expression profiles, the root, stem and leaf were sampled on two-month-old seedlings. Flowers were taken from flowering plants and capsules were obtained during the late seed-producing period. All selected tissues and organs were stored at −80 °C.
Total RNA isolation and quantitative real-time (qRT)-PCR expression analysis
An EasyPure Plant RNA Kit (TransGen Biotech, Beijing, China) was used to extract total RNA according to the manufacturer’s protocol. The first-strand cDNA templates were synthesized from two μg of total RNA, according to the manufacturer’s protocol (Promega, Madison, WI, USA). The reverse transcription reaction was incubated at 42 °C for 65 min in a total volume of 26 μl. The reverse transcription products were diluted five times with sterile ddH2O.
Specific primers were designed by Premier 5.0 according to tobacco EDR1-like gene sequences (Table S1). Due to the high sequence similarity between NtEDR1-1A and NtEDR1-1B, it was difficult to design specific primers for qRT-PCR to measure the expression of NtEDR1-1A and NtEDR1-1B separately. Therefore, one pair of primers targeting both NtEDR1-1A and NtEDR1-1B was synthesized to simultaneously check the expression levels of NtEDR1-1A and NtEDR1-1B. Each PCR reaction was mixed with 10 μl of SYBR Green (TaKaRa, Dalian, China), 6.4 μl of ddH2O, two μl of synthesized cDNA product and 0.8 μl of each primer (50 μM). Relative gene expression level was analyzed according to the 2−ΔΔCt method (Livak & Schmittgen, 2001). Tobacco EF-1a gene (GenBank: D63396.1) was used as the internal reference. The real-time PCR analyses were performed using a qTOWER2.2 real-time PCR system (Analytik Jena AG, Jena, Germany), which was programmed as follows: initial 95 °C denaturation step for 3 min, followed by 40 cycles of denaturation at 95 °C for 10 s and annealing/extension at 60 °C for 1 min (Zhang et al., 2013). Every experiment was conducted with three biological replicates. All data were expressed as mean ± SEM and analyzed by Graphpad Prism 5.0 software.
Results
Identification of the EDR1-like gene family in tobacco and three other plant species
As there was no previous systematic identification and analysis of EDR1-like genes in plants, we identified the EDR1-like genes in tobacco and compared them with other model species. In total, we obtained 19, 12, 8 and 14 EDR1-like genes in tobacco, Arabidopsis, tomato and rice, respectively (Table S2). In addition, the basic sequence information of tobacco EDR1-like genes was analyzed, including the location of the EDR1 regulatory region and the kinase region, the number of amino acids, isoelectric points, molecular weight and predicted location (Table 1).
| Name | Predicted location(s) | Amino acid | Isoelectronic point (pI) | Molecular weight (kDa) | Functional domains (5′–3′) | |
|---|---|---|---|---|---|---|
| EDR1 domain location | Pkinase domain location | |||||
| NtEDR1-1A | Nucleus | 986 | 6.11 | 107.85 | 144–343 | 705–956 |
| NtEDR1-1B | Nucleus | 1,047 | 6.20 | 114.79 | 144–374 | 735–921 |
| NtEDR1-2 | Nucleus | 1,027 | 5.52 | 112.19 | 155–358 | 738–990 |
| NtEDR1-3 | Nucleus | 1,027 | 5.64 | 112.42 | 155–358 | 738–990 |
| NtEDR1-4 | Nucleus | 1,032 | 5.30 | 112.83 | 146–350 | 754–1,006 |
| NtEDR1-5 | Nucleus | 895 | 5.73 | 99.09 | 138–216, 213–313 | 625–877 |
| NtEDR1-6 | Chloroplast, Nucleus | 780 | – | – | 1–83, 92–219 | 501–762 |
| NtEDR1-7 | Nucleus | 846 | 5.74 | 93.33 | 196–423 | 570–824 |
| NtEDR1-8 | Nucleus | 846 | 5.74 | 93.33 | 196–423 | 570–824 |
| NtEDR1-9 | Nucleus | 848 | 5.68 | 93.40 | 200–407 | 572–826 |
| NtEDR1-10 | Nucleus | 776 | 5.38 | 85.15 | 146–353 | 525–774 |
| NtEDR1-11 | Cell membrane | 790 | 5.83 | 89.26 | 55–254 | 534–783 |
| NtEDR1-12 | Nucleus | 891 | 6.25 | 99.48 | 290–497 | 683–890 |
| NtEDR1-13 | Nucleus | 1,240 | 7.56 | 138.74 | 482–681 | 984–1,232 |
| NtEDR1-14 | Cell membrane | 814 | 6.04 | 91.61 | 56–255 | 558–806 |
| NtEDR1-15 | Nucleus | 743 | 5.08 | 81.32 | 146–53 | 525–633, 619–741 |
| NtEDR1-16 | Nucleus | 876 | 6.16 | 98.03 | 288–495 | 681–875 |
| NtEDR1-17 | Cell membrane Nucleus | 648 | 6.36 | 73.67 | 55–166 | 392–641 |
| NtEDR1-18 | Nucleus | 892 | 5.02 | 97.15 | 146–350 | 747–858 |
The NtEDR1 regulatory region was located at the N-terminal, whereas the kinase region was located at the C-terminal of the predicted protein sequence. The C-terminal kinase region shared a sequence identity of 61.48%, and the N-terminal regulatory region had a relatively low identity of 40.52% for the predicted protein sequences of tobacco EDR1-like genes (Fig. S1). The number of amino acids ranged from 648 (NtEDR1-17) to 1,240 (NtEDR1-13). The predicted molecular masses were in the range of 73.67–138.74 kDa and the isoelectric points were 5.02–6.36. Protein subcellular localization prediction is a key step in understanding the protein function and its protein network interaction pattern (Chou & Shen, 2007). In Arabidopsis, at least a fraction of the EDR1 protein has been shown to localize in the nucleus (Christiansen et al., 2011). Our results showed 15 out of 19 tobacco EDR1-like proteins were predicted to be located in the nucleus (Table 1), but NtEDR1-11 and NtEDR1-14 were predicted to be located in the cell membrane. The other two tobacco EDR1-like members, NtEDR1-6 and NtEDR1-17, were shown to be located in the chloroplast or nucleus, and cell membrane or nucleus, respectively. The NLS predictions for most genes were consistent with those of subcellular localization, which showed that 13 NtEDR1-like proteins had NLS sequences (Table 1; Table S3). However, there were differences for another six NtEDR1-like proteins (Table 1; Table S3). This might be due to the different algorithms used for the two tools.
Phylogenetic and analysis of EDR1-like genes
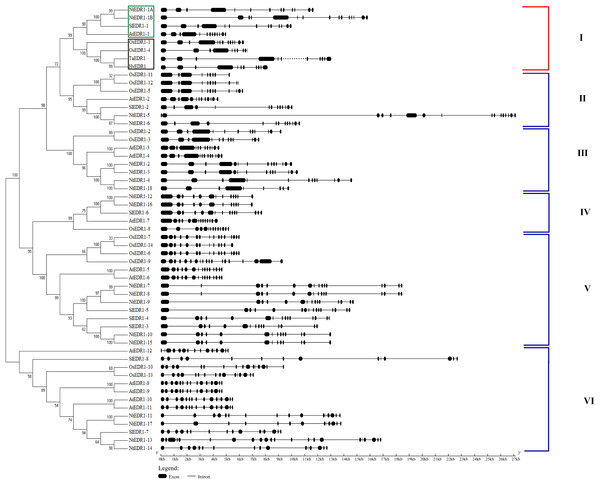
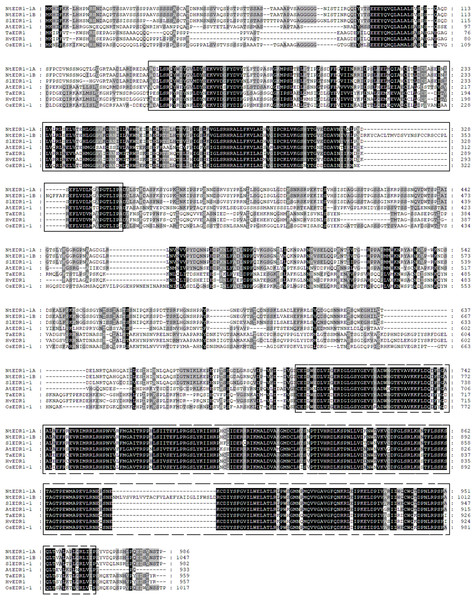
Reports have shown that EDR1 homologs play a negative role in the defense response against powdery mildew in Arabidopsis (Frye & Innes, 1998) and wheat (Zhang et al., 2017). To reveal the evolutionary relationships among different plant species, a phylogenetic tree including EDR1-like gene members in tobacco, Arabidopsis, wheat and rice was constructed using software MEGA 7.0 with NJ method (Fig. 1). The results divided the EDR1-like genes into six clades and the monocot and eudicot EDR1-like genes were separated into two sub-families within each clade (Fig. 1). NtEDR1-1A andNtEDR1-1B, along with EDR1 homologs involved in negative regulation of resistance against powdery mildew in Arabidopsis (AtEDR1-1) and wheat (TaEDR1), were grouped into clade I. Thus, NtEDR1-1A and NtEDR1-1B should be the first choice for further study of any negative roles in tobacco defense against powdery mildew. The exon–intron organizations of the EDR1-like genes were analyzed using online software GSDS (Fig. 1). The exon number of tobacco EDR1-like genes varied from nine (NtEDR1-18) to 22 (NtEDR1-13). Interestingly, NtEDR1-1A, AtEDR1-1, SlEDR1-1 and OsEDR1-1 had 13 exons and were all grouped into clade I in the phylogenetic tree. The similarities of sequences in clade I were further analyzed by multiple sequence alignment. The results showed that both the predicted C-terminal kinase region and N-terminal regulatory region of sequences in clade I had high identities (82.29% and 66.89%, respectively) (Fig. 2).
Figure 1: Phylogenetic relationships and gene structure of EDR1-like gene families from tobacco, Arabidopsis, rice and tomato.
NtEDR1-1A and NtEDR1-1B, along with EDR1 homologs involved in negative regulation of resistance against powdery mildew in Arabidopsis (AtEDR1-1) and wheat (TaEDR1), were grouped into clade I, in which dicotyledon plants are shown with green box, and black box represents monocotyledons. The dotted line represents that there is gap in the available genome sequence of that gene. The GenBank accession number of the barley EDR1 protein sequence is AAG31142.1 (HvEDR1). The genome sequences of barley and wheat were downloaded from the genome databases (ftp://ftp.ensemblgenomes.org/pub/).Figure 2: Protein sequence alignment of EDR1-1 from tobacco, Arabidopsis, rice and tomato.
Positions of EDR1 N-terminal regulatory domains are indicated by a black-lined box; and a dashed box indicates kinase domains.Conserved motif analysis and structure prediction of tobaccoEDR1-like protein
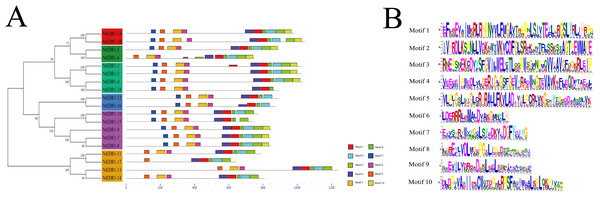
To further analyze the conserved features of the NtEDR1-like family proteins, the motif of the tobacco EDR1-like proteins were predicted by MEME web server. A total of 10 motifs were identified in the NtEDR1-like proteins (Fig. 3; Table S4). According to the prediction of SMART, motifs 1–4 and 10 were located within the kinase domain, while motif 5 was in the EDR1 domain. The results showed motifs 1, 4, 6 and 9 were necessary for all the tobacco EDR1-like members, but motifs 2, 3, 5, 7 and 8 were alternative components (Fig. 3A). The motif logo is shown in Fig. 3B, and the lengths and predicted motif models of each motif are shown in Table S4.
Figure 3: Distribution of conserved motifs in the tobacco EDR1-like family members.
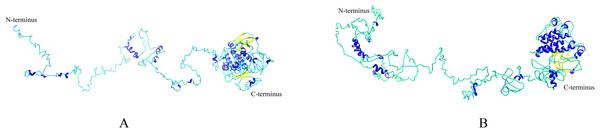
(A) Conserved motifs in NtEDR1-like proteins were analyzed by MEME. Ten different motifs are represented by different colored boxes, and their sizes could be estimated by the scale at the bottom. Details of motif were shown in Table S4. (B) The heights of each box represent the specific amino acid conservation in each motif.Homologous proteins with similar function often have a similar structure. AtEDR1-1 has been experimentally shown to be required for G. cichoracearum susceptibility (Frye & Innes, 1998), so we used AtEDR1-1 as a reference model to analyze NtEDR1-like proteins. The 3D structures of tobacco EDR1-like proteins and AtEDR1-1 were constructed by using I-TASSER web site. The results showed that the predicted 3D model of NtEDR1-1A was similar to that of AtEDR1-1, while the 3D models of the rest tobacco NtEDR1-like proteins were somewhat different (Fig. 4; Fig. S2).
Figure 4: The predicted 3D structures of NtEDR1-1 and AtEDR1-1.
(A) A 3D structure pattern of NtEDR1-1A. (B) A 3D structure pattern of AtEDR1-1. Blue represents α-helices, yellow represents β-strands and cyan represents random coils. Structural images were generated with Chimera 1.2.Expression analysis of EDR1-like genes in tobacco
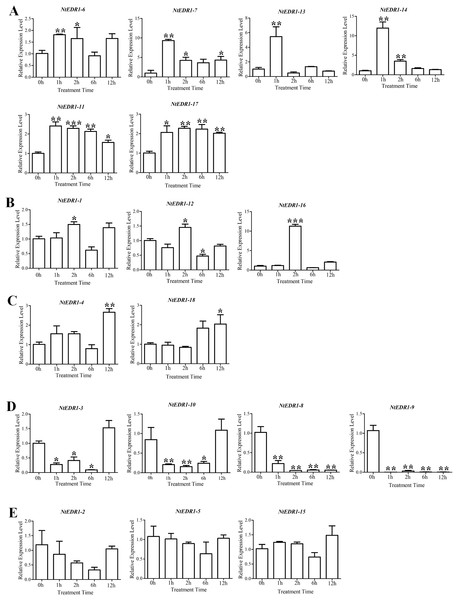
The qRT-PCR was employed to determine the expression of tobacco EDR1-like genes after plants were infected with G. orontii (Fig. 5). NtEDR1-1 was used to represent both NtEDR1-1A and NtEDR1-1B where necessary to describe the transcript abundance of NtEDR1-1A and NtEDR1-1B. The result showed that NtEDR1-1, NtEDR1-12 and NtEDR1-16 had similar expression patterns (Fig. 5B). These genes were not immediately up-regulated but began to increase after 2 hpi. NtEDR1-17 was up-regulated at 1 hpi, and maintained a high level of expression at 2 hpi. NtEDR1-3 and NtEDR1-10 weredown-regulated in the beginning of infection, but up-regulated at 12 hpi. NtEDR1-8 and NtEDR1-9 were significantly down-regulated by G. orontii infection. NtEDR1-2,NtEDR1-5 and NtEDR1-15 showed no obvious changes at all time points tested.NtEDR1-4 and NtEDR1-18 were up-regulated at 12 hpi. NtEDR1-6 and NtEDR1-11 were up-regulated at 1 hpi; subsequently, expression of NtEDR1-6 decreased, and NtEDR1-11 maintained high expression after 2 hpi. NtEDR1-7, NtEDR1-13 and NtEDR1-14 were immediately up-regulated with G. orontii infection at 1 hpi, but their expression declined to a normal level at 6 hpi. These results indicated that different EDR1-like genes showed different expression patterns after G. orontii challenge. The expression patterns of NtEDR1-1, NtEDR1-12 and NtEDR1-16 were similar after tobacco was challenged by G. orontii.
Figure 5: Expression profiles of NtEDR1-like genes in tobacco leaves in response to Golovinomyces orontii infection.
Leaves were inoculated with G. orontii to give approximately 20–25 spores per cm2 and sampled after 0, 1, 2, 12 and 24 h (0 h as control). The expression levels of the genes were significantly increased at 1 h (A), increased at 2 h (B), increased at 12 h (C), decreased (D), and not signifcantly altered (E). ***, ** and * indicate significant differences in comparison with the control at P < 0.001, P < 0.01 and P < 0.05, respectively.Analysis of transcriptional level of NtEDR1-like genes in different tissues/organ samples provides clues on their biological functions. The tissue/organ expression patterns of tobacco NtEDR1-like genes were also investigated using qRT-PCR (Fig. S3). Some NtEDR1-like genes showed organ-specific expression patterns in the organ sample investigated. For example, NtEDR1-13 and NtEDR1-16 were expressed highly only in roots and leaves, respectively (Fig. S3A). The expression of NtEDR1-1, NtEDR1-3, NtEDR1-7, NtEDR1-8, NtEDR1-10 and NtEDR1-15 were absent in flowers; and NtEDR1-4, NtEDR1-9, NtEDR1-11, NtEDR1-14, NtEDR1-17 and NtEDR1-18 did not show any expression in capsules. NtEDR1-2, NtEDR1-5, NtEDR1-6 and NtEDR1-12 had some transcripts in all organ samples tested (Figs. S3B–S3D).
Discussion
The EDR1 genes were first reported to be required for G. cichoracearum susceptibility in Arabidopsis (Frye & Innes, 1998), and are also involved in regulation of multiple physiological activities (Kim et al., 2003). For example, they play a role in the salicylic acid pathway under disease stress, and have a function in the ethylene pathway under drought stress (Rodriguez, Petersen & Mundy, 2010; Tang, Christiansen & Innes, 2005; Tang & Innes, 2002). Moreover, EDR1 could be recruited by EDR4 to the fungal penetration site via physical interaction (Wu et al., 2015). EDR4 has been shown to encode an unknown protein that might function in the same pathway with EDR1 to regulate powdery mildew resistance and cell death (Wu et al., 2015). Therefore, it is necessary to further explore the physiological function of EDR1 members. EDR1s have been found in several plants; however, there were no previous reports on EDR1-like genes in tobacco. In this study, we identified 19 EDR1-like genes in tobacco and compared these with the EDR1-like gene family from Arabidopsis, tomato, and rice. Tissue/organ and G. orontii-induced expression patterns of tobacco EDR1-like genes were further analyzed.
Phylogenetic analysis divided the EDR1-like genes into six clades. NtEDR1-1A and NtEDR1-1B—along with the reported EDR1s, AtEDR1-1 and TaEDR1, negatively functioning in resistance to powdery mildew—clustered in clade I. The results of 3D model prediction showed that NtEDR1-1A was most similar to that of AtEDR1-1 among all the NtEDR1-like genes tested (Fig. 4; Fig. S2). Multi-sequence alignment analysis revealed that genes in clade I showed high similarities, with 82.29% identity of the C-terminal kinase region and 66.89% identity of the N-terminal regulatory region. This suggested that the EDR1 regulatory region and kinase domain were highly conserved in clade I. The pathway of EDR1 homologs is likely conserved between monocots and eudicots (i.e., maize, rice and tomato) (Frye, Tang & Innes, 2001). The EDR1-like members in each clade were divided into monocot and dicot sub-families. Multi-sequence alignment analysis of tobacco EDR1-like proteins showed the C-terminal kinase region had relatively high identity (61.48%), but the N-terminal regulatory region had relatively low identity (40.52%). As EDR1 function is dependent upon the N-terminal regulatory region (Tang, Christiansen & Innes, 2005), the lower identity among the N-terminal regulatory regions of tobacco EDR1-like genes may indicate that they have different functions. Motif distribution analysis showed that different NtEDR1-like members had different numbers and types of motifs (Fig. 3A). Genomic structure analysis also showed that the exon–intron structure of EDR1-like genes greatly differed among species.
Quantitative expressions of tobacco EDR1-like genes following infection by G. orontii were analyzed. The results showed 15 tobacco EDR1-like genes were G. orontii-susceptible (Figs. 5A–5D): NtEDR1-1, NtEDR1-12 and NtEDR1-16 were up-regulated at 2 hpi; NtEDR1-6, NtEDR1-7, NtEDR1-11, NtEDR1-13, NtEDR1-14 and NtEDR1-17 were immediately up-regulated after G. orontii infection; NtEDR1-4 and NtEDR1-18 were up-regulated at 12 hpi; and NtEDR1-3, NtEDR1-8, NtEDR1-9 and NtEDR1-10 were down-regulated at 1 hpi. Knockout of three homologs of TaEDR1 in wheat did not produce complete resistance (Zhang et al., 2017), and separately silencing the tomato homologs of EDR1 (Solyc01 g097980 and Solyc06 g068980) did not result in significant resistance to O. neolycopersici (Gao et al., 2015). It is necessary to silence multiple EDR1 genes to obtain plants with enhanced resistance (Gao et al., 2015). This indicated that multiple EDR1 members may be commonly involved in the negative regulation of resistance against powdery mildew in tobacco and many other plants. The other three NtEDR1-like genes (NtEDR1-2, NtEDR1-5 and NtEDR1-15) did not show significantup- or down-regulation after plant infection by G. orontii (Fig. 5E). Therefore, not all tobacco EDR1-like genes responded to G. orontii infection. Plant basic defenses require the MKK4/MKK5-MPK3/MPK6 kinase cascade, and EDR1 physically associates with MKK4/MKK5 and negatively regulates the MAPK cascade to fine-tune plant innate immunity (Zhao et al., 2014). Several reports have shown that mutations of EDR1 could confer plant disease resistance in Arabidopsis, rice and wheat (Frye & Innes, 1998;Shen et al., 2011; Zhang et al., 2017). Therefore, it might be common that presence of EDR1 may inhibit the plant primary immunity and benefit invasion by powdery mildew.
Tissue/organ expression analysis of tobacco EDR1-like genes was carried out usingqRT-PCR. The results showed that different EDR1-like genes had different expression patterns. NtEDR1-2, NtEDR1-3, NtEDR1-4, NtEDR1-9 and NtEDR1-10 had higher expression levels in roots. NtEDR1-1, NtEDR1-5, NtEDR1-7, NtEDR1-8, NtEDR1-12 and NtEDR1-15 were predominantly expressed in capsules. NtEDR1-11 and NtEDR1-17 were highly expressed in flowers. Interestingly, two NtEDR1-like genes, NtEDR1-13 and NtEDR1-16, were exclusively expressed in roots and leaves, respectively. In addition to functions as negative regulators of plant defense, EDR1-like genes have also been shown to have roles in other physiological processes. For example, OsEDR1-1 (Os3 g06410) had a high expression level during maturation of the panicle before heading, after heading and at maturity (pollination stage) in rice, and was further suggested to play a role in plant growth and development, and in maturity of panicles (Kim et al., 2003). In our study, NtEDR1-11 and NtEDR1-17 were mainly expressed in flowers, whereas NtEDR1-5, NtEDR1-7,NtEDR1-8, NtEDR1-12 and NtEDR1-15 were highly expressed in capsules. Thus, some tobacco EDR1-like genes may also have a physiological function during the plant reproductive period.
Conclusions
In this research, 19 EDR1-like genes were identified in a genome-wide analysis in N. tabacum. Through multiple sequences alignment, phylogenetic analysis, gene structure analysis and comparative analysis of predicted 3D structures, NtEDR1-1A was shown to have the highest similarity to AtEDR1-1, which has been reported to be responsive to powdery mildew. Expression profiles revealed that NtEDR1-1 (NtEDR1-1A and NtEDR1-1B) was not the only tobacco EDR1-like member responsive to G. orontii, but not all tobacco EDR1-like genes were responsive to G. orontii. Tissue/organ expression profiles showed that different EDR1-like members had different expression patterns, but some NtEDR1-like genes showed tissue-specific expression patterns. Our results provide foundational information on the EDR1-like gene family in tobacco species, and promise to promote future research on the functions of EDR1-like genes in plants.
Supplemental Information
Multiple sequences alignment of the protein sequences of tobacco EDR1-like genes.
Expression profile of tobacco EDR1-like genes in different tissue/organ samples.
Root-Y, Stem-Y and Leaf-Y indicate young root, young stem and young leaf, respectively. Flower-F indicates open flowers. Capsule-seed indicates the capsules that were obtained during the late seed-breeding period. The genes have clear tissue-specific expression patterns (A); The genes expressed in all organ samples tested (B); The expression of genes were absent in flowers (C); and the genes did not show any expression in capsules (D).
The primer sequences used in qRT-PCR for detection the expression patterns of NtEDR1-like genes.
Summary of the EDR1-like genes in different plant species.
The protein sequences of EDR1-like genes in each species is presented in Supplementary Materials (Arabidopsis in File S1; tobacco in File S2; tomato in File S3; and rice in File S4).
Prediction of NLS sequences and the existence of transmembrane helices of the tobacco EDR1-like proteins.
“√” represents protein has transmembrane helices.