Comparison of Symbiodiniaceae diversities in different members of a Palythoa species complex (Cnidaria: Anthozoa: Zoantharia)—implications for ecological adaptations to different microhabitats
- Published
- Accepted
- Received
- Academic Editor
- Konstantinos Kormas
- Subject Areas
- Biodiversity, Marine Biology, Molecular Biology
- Keywords
- Zoantharia, Palythoa species complex, Symbiodiniaceae, Ecological divergence
- Copyright
- © 2020 Mizuyama et al.
- Licence
- This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, reproduction and adaptation in any medium and for any purpose provided that it is properly attributed. For attribution, the original author(s), title, publication source (PeerJ) and either DOI or URL of the article must be cited.
- Cite this article
- 2020. Comparison of Symbiodiniaceae diversities in different members of a Palythoa species complex (Cnidaria: Anthozoa: Zoantharia)—implications for ecological adaptations to different microhabitats. PeerJ 8:e8449 https://doi.org/10.7717/peerj.8449
Abstract
In this study we compared genotypes of zoantharian host-associating algal symbionts among Palythoa species, which are among the dominant benthic reef organisms in the Ryukyu Archipelago, Japan, and evaluated Symbiodiniaceae diversities of closely related congeneric Palythoa species. We targeted a species complex of the zoantharian genus Palythoa (P. tuberculosa, P. sp. yoron, P. mutuki) living among different microhabitats in a narrow reef area of Tokunoshima Island. For phylogenetic analyses, we used two DNA marker regions; nuclear internal transcribed spacer (ITS) and plastid mini-circle non-coding region (psbAncr), both of which have previously been used to determine Symbiodiniaceae genotypes of zoantharian species. Our results showed that all Palythoa species hosted symbionts of the genus Cladocopium, with genotypic compositions of this genus showing some variations among the three different Palythoa species. Additionally, we found that the Cladocopium genotypic composition was statistically different among Palythoa species, and among P. tuberculosa specimens in different microhabitats. Our results suggest that ecological divergence among these three Palythoa species may be related to differing Symbiodiniaceae diversities that may in turn contribute to eco-physiological adaptation into different microhabitats on coral reefs.
Introduction
Zoantharians (Anthozoa: Zoantharia) belong to the phylum Cnidaria and can be dominant organisms in shallow coral reef areas (e.g., Burnett et al., 1994). In particular, the genus Palythoa is often among the most dominant benthos in coral reef areas (Irei, Nozawa & Reimer, 2011; Santos et al., 2016; Reimer et al., 2017a).
We recently reported on four putative Palythoa species (P. tuberculosa, P. sp. yoron, P. mutuki, and P. aff. mutuki) that form a species complex, and were observed to all occur within a narrow range of coral reefs in southern Japan (Mizuyama, Masucci & Reimer, 2018). For example, P. tuberculosa tends to occur across a wide range of habitats from shallow to deeper areas, from the intertidal zone to the mesophotic reef slope (Mizuyama, Masucci & Reimer, 2018), and has been reported from tropical to temperate regions (Reimer, Takishita & Maruyama, 2006). On the other hand, the other three Palythoa species appear to more restricted compared to P. tuberculosa in terms of their distribution and habitats within coral reefs. Palythoa mutuki is the second most dominant species in this genus in Okinawa and is often dominant at the reef edge, in surge channels, and in small bumps on reef flats (Irei, Nozawa & Reimer, 2011). Palythoa sp. yoron has yet to be formally described, but tends to occur on reef flats and backreef moats where it is exposed to strong water currents (Shiroma & Reimer, 2010). Although there is little published information on P. aff. mutuki, it has been observed near P. mutuki colonies on the reef flat (Mizuyama, Masucci & Reimer, 2018). Although molecular delineation of these Palythoa species groups was unsuccessful with molecular data, likely due to incomplete lineage sorting, they can be distinguished via morphological and reproductive data (Mizuyama, Masucci & Reimer, 2018). In addition, these Palythoa species display different microhabitat patterns within the coral reef, but it is still unclear how these species would have diversified under almost completely sympatric conditions.
Symbiodiniaceae endosymbiotic dinoflagellates are symbiotic with various metazoan phyla including Cnidaria (LaJeunesse et al., 2018). Many zoantharians maintain Symbiodiniaceae, similar to reef-building corals (Noda et al., 2017; Wee, Kurihara & Reimer, 2019). In the case of scleractinian corals, symbiotic relationships with Symbiodiniaceae are important for host survival in various environments (Baker, 2003), and can contribute to ecological divergence of coral host species (Winters et al., 2009). Previous molecular studies have reported that species composition of Symbiodiniaceae is closely related to host genotypes in corals (e.g., Bongaerts et al., 2010; Pinzon & LaJeunesse, 2011). Thus, information on the composition Symbiodiniaceae of the four Palythoa species above would also be helpful to understand their ecological divergence into different microenvironments within a reef. In particular, genotypic composition of symbiotic algae would be informative for understanding ecological divergence of these species because the genetic and/or community changes of microbiomes are expected to be faster than that of the hosts themselves (Torda et al., 2017), facilitating eco-physiological adaptation of holobionts into different microenvironments (e.g., Reimer et al., 2017b; Wee, Kurihara & Reimer, 2019). In this study, we aimed to (1) compare diversities of symbionts among the closely related Palythoa species P. tuberculosa, P. sp. yoron, P. mutuki and P. aff. mutuki, and (2) determine if diversities of symbionts explain eco-physiological adaptations to microhabitats of each species that entailed divergences among them (P. tuberculosa, P. sp. yoron and P. mutuki).
Materials & Methods
Specimens collection
Eighty-two colonies of three Palythoa species (P. tuberculosa, P. sp. yoron, and P. mutuki) were collected from a shallow fringing reef of Tokunoshima Island, Kagoshima, Japan (Figs. 1 and 2). Specimens of these three Palythoa species were collected in four different areas (Table 1, Fig. 2A): reef edge (Fig. 2B, 27.76998333N, 129.03988611E) for P. tuberculosa (Fig. 2C); reef flat 1 (Fig. 2D, 27.76997777N, 129.03925000E) for P. tuberculosa (Fig. 2E) and P. mutuki; reef flat 2 (Fig. 2F, 27.77195277N, 129.03843611E) for P. mutuki (Fig. 2G); and backreef moat (Fig. 2H, 27.76990833N, 129.03855833E) for P. tuberculosa and P. sp. yoron (Fig. 2I). To avoid collecting clones, we collected individuals from clearly different colonies while maintaining a set distance from each other of at least 1 m. In a previous study, even when closer to each other (within approximately 50 × 50 cm), no clones were observed in Zoanthus (Cnidaria: Anthozoa: Zoantharia) colonies (Albinsky et al., 2018). In addition, eighteen previously collected specimens of Palythoa species including 10 P. aff. mutuki specimens from Mizuyama, Masucci & Reimer (2018) were also examined in this study (Table 1).
Figure 1: Location of Tokunoshima Island and the sampling site (arrow in inset) for the Palythoa specimens in this study.
Map data: GeoLite2 data created by MaxMind using the Generic Mapping Tools (GMT v5.4.5) software package. CC BY SA 4.0.Figure 2: Landscape of the coral reef flat at the study site and in situ images of Palythoa species used in this study.
(A) Satellite image of the reef area obtained by Google Earth; (B) reef edge; (C) P. tubeculosa; (D) reef flat 1; (E) P. tuberculosa; (F) reef flat 2; (G) P. mutuki; (H) backreef moat; (I) P. sp. yoron. Map data: Google, Maxar Technologies. Scale bars in C, E, G, and I are 10 cm.DNA extraction and PCR amplification
From each of these specimens, several polyps were cut with a surgical knife and DNA was extracted using DNeasy Blood and Tissue Kit (QIAGEN). DNA concentrations were checked by Qubit Fluorometer (ThermoFisher, Waltham, USA). Two molecular markers for genotyping symbiotic algae of Palythoa species were examined: nuclear internal transcribed spacer ribosomal DNA (ITS-rDNA) region including partial 18S–ITS1–5.8S–ITS2–partial 28S (primers zITSf: CCG GTG AAT TAT TCG GAC TGA CGC AGT and ITS4: TCC TCC GCT TAT TGA TAT GC, (Baillie, Belda-Baillie & Maruyama, 2000; appx. 700–750 bp) and plastid mini-circle non-coding region DNA (psbAncr) (primers 7.4-Forw: GCA TGA AAG AAA TGC ACA CAA CTT CCC and 7.8-Rev: GGT TCT CTT ATT CCA TCA ATA TCT ACT G, (Moore et al., 2003; appx. 800–850 bp). These regions were amplified according to the PCR thermal conditions in Wee, Kurihara & Reimer (2019). Amplified PCR products of symbionts were directly sequenced, and sequence data were manually checked based on the chromatogram files and low quality sites were removed at the 5′ and 3′ ends by BioEdit v.7.0.5.3 (Hall, 1999). Obtained sequences were deposited in the GenBank database (MN654128–MN654306, Table 1).
| Specimen ID | Location/Region | Spiecies ID | Date (m/d/y) | Environment | Accession no. of ITS | Accession no. of psbA-F | Accession no. of psbA-R |
|---|---|---|---|---|---|---|---|
| A01PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef edge | MN654209 | MN654185 | – |
| A02PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef edge | MN654210 | MN654184 | MN654134 |
| A03PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef edge | MN654211 | – | – |
| A04PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef edge | MN654212 | MN654186 | MN654135 |
| A05PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef edge | MN654213 | MN654187 | MN654136 |
| A06PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef edge | MN654214 | MN654188 | – |
| A07PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef edge | MN654215 | MN654189 | MN654137 |
| A08PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef edge | MN654216 | MN654190 | MN654138 |
| A09PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef edge | MN654217 | – | – |
| A11PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef flat | MN654218 | MN654191 | MN654139 |
| A12PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef flat | MN654219 | MN654192 | MN654140 |
| A13PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef flat | MN654220 | MN654193 | – |
| A14PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef flat | MN654221 | – | – |
| A15PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef flat | MN654222 | – | – |
| A16PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef flat | MN654223 | MN654194 | MN654141 |
| A17PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef flat | MN654224 | – | – |
| A18PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef flat | MN654225 | MN654195 | MN654142 |
| A19PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef flat | MN654226 | MN654198 | – |
| A20PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Reef flat | MN654227 | – | – |
| A21PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Backreef moat | MN654228 | MN654169 | MN654159 |
| A22PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Backreef moat | MN654229 | – | – |
| A24PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Backreef moat | MN654230 | – | MN654143 |
| A25PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Backreef moat | MN654231 | – | – |
| A26PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Backreef moat | MN654232 | – | – |
| A27PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Backreef moat | MN654233 | – | – |
| A28PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Backreef moat | MN654234 | – | – |
| A29PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Backreef moat | MN654235 | – | – |
| A30PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Jun 2, 2019 | Backreef moat | MN654236 | – | – |
| B01PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 2, 2019 | Reef flat | MN654237 | – | – |
| B02PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 2, 2019 | Reef flat | MN654238 | MN654199 | MN654144 |
| B03PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 2, 2019 | Reef flat | MN654239 | – | – |
| B04PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 2, 2019 | Reef flat | MN654240 | – | – |
| B05PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 2, 2019 | Reef flat | MN654241 | – | MN654145 |
| B06PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 2, 2019 | Reef flat | MN654242 | MN654170 | MN654160 |
| B07PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 2, 2019 | Reef flat | MN654243 | – | MN654161 |
| B08PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 2, 2019 | Reef flat | MN654244 | MN654171 | MN654162 |
| B09PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 2, 2019 | Reef flat | MN654245 | – | – |
| B11PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 2, 2019 | Reef flat | MN654246 | – | – |
| B12PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | MN654247 | MN654172 | – |
| B13PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | MN654248 | – | – |
| B14PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | MN654249 | MN654173 | – |
| B15PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | MN654250 | – | – |
| B16PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | MN654251 | – | – |
| B17PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | MN654252 | MN654174 | MN654163 |
| B18PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | MN654253 | MN654200 | – |
| B20PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | MN654254 | – | – |
| B21PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | MN654255 | – | – |
| B22PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | MN654256 | – | – |
| B23PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | MN654257 | – | – |
| B24PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | MN654258 | MN654175 | MN654164 |
| B25PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | – | MN654176 | MN654165 |
| B26PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | MN654259 | – | MN654166 |
| B28PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Jun 3, 2019 | Reef flat | – | MN654177 | MN654167 |
| C01PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654260 | – | – |
| C02PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654261 | – | – |
| C03PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654262 | – | – |
| C04PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654263 | – | – |
| C05PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654264 | – | – |
| C06PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654265 | – | – |
| C07PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654266 | – | – |
| C08PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654267 | – | – |
| C09PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654268 | – | – |
| C10PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654269 | – | – |
| C11PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654270 | – | – |
| C12PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654271 | MN654201 | MN654146 |
| C13PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654272 | – | – |
| C14PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654273 | MN654179 | MN654147 |
| C15PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654274 | MN654180 | – |
| C16PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654275 | MN654202 | MN654148 |
| C17PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654276 | MN654203 | MN654149 |
| C18PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 2, 2019 | Backreef moat | MN654277 | – | MN654168 |
| C19PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 3, 2019 | Backreef moat | MN654278 | – | – |
| C20PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 3, 2019 | Backreef moat | MN654279 | MN654204 | MN654150 |
| C21PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 3, 2019 | Backreef moat | MN654280 | MN654205 | MN654151 |
| C22PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 3, 2019 | Backreef moat | MN654281 | MN654206 | MN654152 |
| C24PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 3, 2019 | Backreef moat | MN654282 | MN654181 | – |
| C25PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 3, 2019 | Backreef moat | MN654283 | MN654196 | MN654153 |
| C26PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 3, 2019 | Backreef moat | MN654284 | – | MN654154 |
| C27PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 3, 2019 | Backreef moat | MN654285 | MN654207 | MN654155 |
| C28PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 3, 2019 | Backreef moat | MN654286 | – | – |
| C29PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 3, 2019 | Backreef moat | MN654287 | MN654208 | MN654156 |
| C30PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Jun 3, 2019 | Backreef moat | MN654288 | MN654182 | MN654157 |
| 159PamToKa | Kaminomine/Tokunoshima | Palythoa aff. mutuki | July 28, 2010 | In Mizuyama, Masucci & Reimer (2018) | MN654300 | – | – |
| 233PamErYa | Yakomo/Okinoerabu | Palythoa aff. mutuki | Jun 17, 2011 | In Mizuyama, Masucci & Reimer (2018) | MN654301 | – | – |
| 237PamErSu | Sumiyoshi/Okinoerabu | Palythoa aff. mutuki | Jun 18, 2011 | In Mizuyama, Masucci & Reimer (2018) | MN654302 | – | – |
| 248PamToKa | Kaminomine/Tokunoshima | Palythoa aff. mutuki | Jun 21, 2011 | In Mizuyama, Masucci & Reimer (2018) | MN654303 | – | – |
| 250PamToKa | Kaminomine/Tokunoshima | Palythoa aff. mutuki | Jun 21, 2011 | In Mizuyama, Masucci & Reimer (2018) | MN654304 | MN654183 | MN654131 |
| 328PamOkTe | Teniya/Okinawa | Palythoa aff. mutuki | Apr 5, 2012 | In Mizuyama, Masucci & Reimer (2018) | MN654305 | – | – |
| 364PamOkOk | Oku/Okinawa | Palythoa aff. mutuki | Jun 25, 2012 | In Mizuyama, Masucci & Reimer (2018) | MN654306 | – | – |
| 2PtOkOd | Odo/Okinawa | Palythoa tuberculosa | Aug 18, 2009 | In Mizuyama, Masucci & Reimer (2018) | MN654289 | – | MN654158 |
| 39PtYoUk | Ukachi/Yoron | Palythoa tuberculosa | Mar 4, 2010 | In Mizuyama, Masucci & Reimer (2018) | MN654290 | – | MN654132 |
| 63PtErYa | Yakomo/Okinoerabu | Palythoa tuberculosa | Mar 5, 2010 | In Mizuyama, Masucci & Reimer (2018) | MN654291 | – | MN654133 |
| 100PtToKa | Kaminomine/Tokunoshima | Palythoa tuberculosa | Mar 9, 2010 | In Mizuyama, Masucci & Reimer (2018) | MN654292 | – | MN654128 |
| 15PyOkOd | Odo/Okinawa | Palythoa sp. yoron | Sep 5, 2009 | In Mizuyama, Masucci & Reimer (2018) | MN654297 | – | MN654130 |
| 51PyYoUk | Ukachi(West)/Yoron | Palythoa sp. yoron | Mar 4, 2010 | In Mizuyama, Masucci & Reimer (2018) | MN654298 | – | – |
| 85PyErYa | Yakomo/Okinoerabu | Palythoa sp. yoron | Mar 5, 2010 | In Mizuyama, Masucci & Reimer (2018) | MN654296 | MN654197 | – |
| 105PyToKa | Kaminomine/Tokunoshima | Palythoa sp. yoron | Mar 9, 2010 | In Mizuyama, Masucci & Reimer (2018) | MN654299 | MN654178 | MN654129 |
| 218PmOkOd | Odo/Okinawa | Palythoa mutuki | May 4, 2011 | In Mizuyama, Masucci & Reimer (2018) | MN654294 | – | – |
| 77PmErYa | Yakomo/Okinoerabu | Palythoa mutuki | Mar 5, 2010 | In Mizuyama, Masucci & Reimer (2018) | MN654293 | – | – |
| 280PmToKa | Kaminomine/Tokunoshima | Palythoa mutuki | Oct 5, 2011 | In Mizuyama, Masucci & Reimer (2018) | MN654295 | – | – |
Haplotype network inference and phylogenetic estimation
Obtained sequences for ITS-rDNA, psbAncr forward and reverse regions were aligned, respectively. In order to discriminate taxa of Symbiodiniaceae, we extracted the ITS2 region utilizing SymPortal (Hume et al., 2019; https://symportal.org/) and performed BLASTN search against the nt database using the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi) for ITS-rDNA sequences. Haplotype network inference was performed for ITS-rDNA sequences using the alignment with TCS networks method (Clement et al., 2002) in PopART (Leigh & Bryant, 2015). Any columns in the alignment with gaps or ambiguous sites were automatically masked in the inference. The phylogenetic analyses were performed by MEGA version X (Kumar et al., 2018) and any loci with ambiguous (double peaks) sites and gaps was automatically deleted completely for calculation in order to avoid over/underestimation of genetic distance among each sequence. Molecular phylogenetic trees of each marker were constructed by maximum likelihood (ML) and neighbor joining (NJ) methods under the JC+G model for ITS-rDNA region and the JC model for psbAncr regions adopted by modeltest program within MEGA X. The significance of each node was tested by bootstrap test with 1,000 replications. Bayesian inference was performed using BEAST2 (Bouckaert et al., 2019) under default settings other than the clock model being changed to the relaxed log normal model, which showed the highest likelihood value according to the model comparison program compiled in BEAST2 (Drummond et al., 2006). Posterior probability (PP) on each branch was calculated summarizing four independent 10 million MCMC simulations.
Statistical analyses
To clarify the relationships between (1) symbiont lineages and host species, and (2) symbiont lineages and host microhabitats, Fisher’s exact test was conducted for the compositions of genotype for ITS-rDNA region and monophyletic clades for psbAncr forward and reverse regions. It should be noted that host microhabitat was restricted by host species for P. sp. yoron and P. mutuki, and thus we only targeted P. tuberculosa for these analyses (aim 2 above) When significance was detected in Fisher’s exact test, Cramér’s coefficient of association (V) was calculated to evaluate which factors (host species or host microhabitat) were strongly associated with each other.
Results
Sequence alignment
The total number of sequences of Symbiodiniaceae from specimens of the four Palythoa species obtained in this study was 98 sequences for the ITS-rDNA region (513–773 bp), 40 sequences for the psbAncr forward region (330–547 bp), and 41 sequences for the psbAncr reverse region (352–494 bp). As the primer set for psbAncr used in this study did not make a congruent contig, obtained sequences of forward regions and reverse regions were aligned separately (Noda et al., 2017). After alignment, a total of 449 sites with 5 parsimony informative (=PI) sites for the ITS-rDNA region, 260 sites with 94 PI sites for the psbAncr forward region, and 293 sites with 40 PI sites for the psbAncr reverse region were used for each phylogenetic estimation.
Barcoding, haplotype network and phylogenetic trees
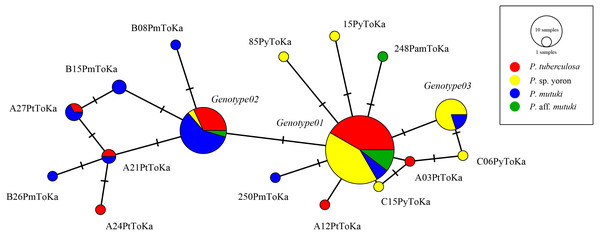
As the result of BLAST searches, all query sequences of the ITS-rDNA region (n = 98) were confirmed as belonging to the genus Cladocopium. Seventeen ITS-rDNA unique sequences (=genotypes) were observed in TCS network, with most of the sequences belonging to one of major three ITS-rDNA genotypes (Fig. 3, Table S1). No significant clade was detected for the ITS-rDNA phylogenetic tree (Fig. S1). Summarizing these ITS-rDNA genotypes from the viewpoint of host species, P. tuberculosa possessed mainly Genotype01 (n = 20) followed by Genotype02 (n = 7), and P. sp. yoron also possessed mainly Genotype01 (n = 20) followed by Genotype03 (n = 8) (see details in Table S1). On the other hand, P. mutuki possessed mainly Genotype02 (n = 13) with a few Genotype01 (n = 3) and Genotype03 (n = 2). Although the number of specimens examined was smaller (n = 6) than those the other species, P. aff. mutuki also possessed mainly Genotype01 (n = 5).
Figure 3: Haplotype network tree constructed with nuclear ITS-rDNA region alignment using TCS networks method.
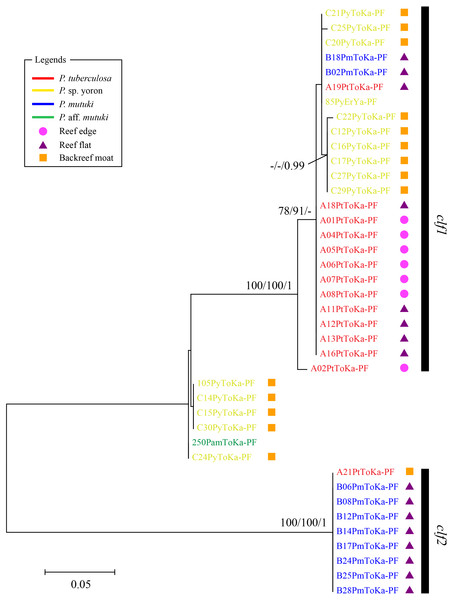
Scale represents number of sequences with circle sizes proportional to haplotype frequency. Colors represent Palythoa species: red, P. tuberculosa; yellow, P. sp. yoron; blue, P. mutuki; green, P. aff. mutuki.In contrast, phylogenetic trees generated from psbAncr regions had a higher resolution. Two monophyletic clades were well supported by bootstrap values and posterior probability in both forward (Fig. 4 clf1, ML = 100, NJ = 100, PP = 1 and clf2, ML = 100, NJ = 100, PP = 1) and reverse trees (Fig. 5 clr1 and clr2, ML = 100, NJ = 100, PP = 1). Summarizing these Symbiodiniaceae lineages from the viewpoint of host species, P. tuberculosa inhabiting the reef edge possessed clf1/clr1 lineage (n = 7∕5) and one specimen inhabiting at the backreef moat possessed clf2/clr2 lineage. Palythoa sp. yoron inhabiting at the backreef moat possessed mainly clf1/clr1 (n = 9∕13), however, approximately one third of specimens (n = 5) possessed other lineages. On the other hand, P. mutuki inhabiting the reef flat possessed mainly clf2/clr2 (n = 8∕8) other than two specimens that possessed clf1/clr1. Unfortunately, as most of P. aff. mutuki specimens were not amplified by this primer set, we could only obtain phylogenetic information on one specimen which possessed the same lineage as P. sp. yoron (C24ToKa-PF) for the forward region and clr1 for the reverse region.
Figure 4: Molecular phylogenetic tree of Symbiodiniaceae of Palythoa species using mitochondrial psbAncr forward region.
Bootstrap values of maximum likelihood (ML) and neighbor joining (NJ) methods, and posterior probability (PP) are shown more than 70% for ML and NJ, and more than 0.95 for PP at the nodes, respectively. Scale bars indicate substitutions per site. Colored letters and colored diagrams represent Palythoa species and their habitats, respectively: red, P. tuberculosa; yellow, P. sp. yoron; blue, P. mutuki; green, P. aff. mutuki; circle in pink, reef edge; triangle in purple, reef flat; square in orange, backreef moat.Figure 5: Molecular phylogenetic tree of Symbiodiniaceae of Palythoa species using mitochondrial psbAncr reverse region.
Bootstrap values of maximum likelihood (ML) and neighbor joining (NJ) methods, and posterior probability (PP) are shown more than 70% for ML and NJ, and more than 0.95 for PP at the nodes, respectively. Scale bars indicate substitutions per site. Colored letters and colored diagrams represent Palythoa species and their habitats, respectively: red, P. tuberculosa; yellow, P. sp. yoron; blue, P. mutuki; green, P. aff. mutuki; circle in pink, reef edge; triangle in purple, reef flat; square in orange, backreef moat.Relationships among symbiont genotype/lineages, host species and host microhabitats
From the results of Fisher’s Exact test, significant differences were detected in all combinations, i.e., ITS-rDNA genotype and host species (p < 0.01), psbAncr forward lineages and host species (p < 0.01), psbAncr reverse lineages and host species (p < 0.01), and ITS-rDNA genotype and host microhabitats for P. tuberculosa (p < 0.05) (Table 2). In other words, it was shown that Symbiodiniaceae lineages and host species were not independent, nor were Symbiodiniaceae lineages and host microhabitats for P. tuberculosa. The effective dose calculated by Cramér’s coefficient of association (V) was largest between host species and psbAncr forward/reverse lineages (V = 0.786, V = 0.682, respectively), and moderate for the other combinations (host species and ITS-rDNA genotypes, V = 0.477; host microhabitats and ITS-rDNA genotypes).
| Symbiodiniaceae genotype (ITS-rDNA) | Symbiodiniaceae lineage (psbAncr forward region) | Symbiodiniaceae lineage (psbAncr reverse region) | ||||||
|---|---|---|---|---|---|---|---|---|
| Genotype01 | Genotype02 | Genotype03 | clf1 | clf2 | clr1 | clr2 | ||
| Host species | P. tuberculosa | 20 | 7 | 0 | 13 | 1 | 13 | 2 |
| P. sp. yoron | 20 | 1 | 8 | 10 | 0 | 14 | 1 | |
| P. mutuki | 3 | 13 | 2 | 2 | 8 | 2 | 8 | |
| P. aff. mutuki | 5 | 1 | 0 | – | – | – | – | |
| Total | 48 | 22 | 10 | 25 | 9 | 29 | 11 | |
| p < 0.01, V = 0.477 | p < 0.01, V = 0.786 | p < 0.01, V = 0.682 | ||||||
| Host habitats of P. tuberculosa | Reef edge | 8 | 0 | |||||
| Reef flat | 7 | 2 | ||||||
| Backreef moat | 2 | 4 | ||||||
| Total | 17 | 6 | ||||||
| p < 0.05, V = 0.508 | ||||||||
Notes:
P. aff. mutuki was removed from statistical analyses of psbAncr region due to low numbers of specimens.
Discussion
Symbiodiniaceae genotype/lineage and host species
The development of molecular markers such as psbAncr that have higher resolution than commonly used 18S or ITS ribosomal DNA markers has helped unveil a more detailed picture of the genetic diversity of Symbiodiniaceae (Takishita et al., 2003; LaJeunesse & Thornhill, 2011; LaJeunesse et al., 2018) (but see also Hume et al., 2019 who utilized intragenomic variation of ITS2 to resolve genetic delineations). Accordingly, host species biodiversity has been discovered from the initial observation of differences of Symbiodiniaceae phylotypes in some cnidarian species (e.g., gorgonian Eunicea flexuosa, Prada et al., 2014; scleractinian coral Seriatopora hystrix, Warner, Van Oppen & Willis, 2015).
From the results of Mizuyama, Masucci & Reimer (2018), none of the four molecular markers utilized could clearly delineate four Palythoa species, although they could delineate two closely related species groups composed of P. tuberculosa—P. sp. yoron and P. mutuki—P. aff. mutuki. These previous results seem to be reflected in the results in the current study of Symbiodiniaceae genotypes of ITS-rDNA and lineages of psbAncr regions. Palythoa tuberculosa and P. sp. yoron mostly shared the same symbiont genotype (Genotype01); nevertheless, they also partially shared the other genotypes with P. mutuki (Genotype02 and Genotype03). With regard to psbAncr lineages, even though the delineation of species groups between P. tuberculosa—P. sp. yoron and P. mutuki were shown more clearly, they were not divided completely. The situation requires further investigation via obtaining more P. aff. mutuki specimens’ psbAncr sequences. Unfortunately, in the current study, despite much searching, we could not find large numbers of P. aff. mutuki on the reef in Tokunoshima Island, even though they were previous sampled for Mizuyama, Masucci & Reimer (2018). We do not know what happened to P. aff. mutuki colonies, but they may have been strongly affected by the bleaching events of 2016 and 2017 observed in southern Japan (Masucci et al., 2019).
Symbiodiniaceae genotype/lineage and microhabitat of host species
From the results of the phylogenetic analyses, three microhabitats were not exclusively allocated in distinct Symbiodiniaceae genotypes or monophyletic clades, but the ratios of different genotypes were significantly different for P. tuberculosa. Regarding P. tuberculosa, Symbiodiniaceae Genotype01 was mostly detected on the reef edge and reef flat, while Genotype02 was mainly observed in the backreef moat. Although there were not enough samples to conduct statistical examinations of P. sp. yoron and P. mutuki due to their habitat specificity, Genotype02 and clf2/clr2 were detected mainly on the reef flat, while Genotype01 and clf1/clr1 were observed from all three environments.
It has been reported that zoantharian species with different symbiotic genotypes show species-specific photosynthetic responses against seawater temperature and p CO2 (Graham & Sanders, 2016; Reimer et al., 2017b; Wee, Kurihara & Reimer, 2019). Although the four Palythoa species in this study occurred sympatrically on one reef, the environmental conditions in a reef can be quite different according to small-scale geographical features. Seawater temperatures on reef flats frequently reach near 40 °C (Achituv & Dubinsky, 1990). In enclosed reefs, seawater temperatures and p CO2 show higher variations than those in exposed reefs (Suzuki, Nakamori & Kayanne, 1995; Fitt et al., 2001). Thus, the relationship between Symbiodiniaceae and host Palythoa species may change among different microhabitats in a reef area, facilitating ecological divergence of Palythoa species within a narrow geographic range.
Although a previous molecular study could not distinguish the boundaries among these Palythoa species (Mizuyama, Masucci & Reimer, 2018), it is suggested by our results that these species are ecologically divergent, and physiological differences within Symbiodiniaceae species may contribute to their ecological adaptation. In fact, Howells et al. (2012) reported that Cladocopium C1 in Acropora tenuis showed different physiological responses between northern and southern populations in the Great Barrier Reef. Considering that Cladocopium contains various species distinguished by differences of only a few bp in the ITS2 maker (Thornhill et al., 2014), meta-barcoding analyses via next-generation sequencing would be necessary to further understand the detailed relationship between Symbiodiniaceae and Palythoa species complex.
Conclusions
We succeeded in obtaining genotypic data of Symbiodiniaceae from four putative Palythoa species and detected micro-scale geographic variations of the symbiotic algae among these species within a single coral reef. Our results suggest that ecological divergence among Palythoa species may be related to differences in Symbiodiniaceae diversities among microhabitats, even within a narrow reef area. More powerful genetic data such as that generated by next-generation sequencing could provide us with additional understanding on how neighboring Palythoa species have co-evolved with Symbiodiniaceae among the different microhabitats in a reef.
Supplemental Information
Composition of genotypes for ITS-rDNA sequences of Simbiodiniaceae from 4 Palythoa species
Molecular phylogenetic tree of Symbiodiniaceae of Palythoa species using sequences of the nuclear ITS-rDNA region
Shaded boxes represent three main genotypes occupying most sequences from four Palythoa species. Bootstrap values of maximum likelihood (ML) and neighbor joining (NJ) methods, and posterior probability (PP) are shown more than 70% for ML and NJ, and more than 0.95 for PP at the nodes, respectively. Scale bars indicate substitutions per site. Colored letters and colored diagrams represent Palythoa species and their habitats, respectively: red, P. tuberculosa; yellow, P. sp. yoron; blue, P. mutuki; green, P. aff. mutuki; circle in pink, reef edge; triangle in purple, reef flat; square in orange, backreef moat.